Figure 2. Robust induction of TXNIP requires activation of IRE1α’s bifunctional kinase and RNase domains.
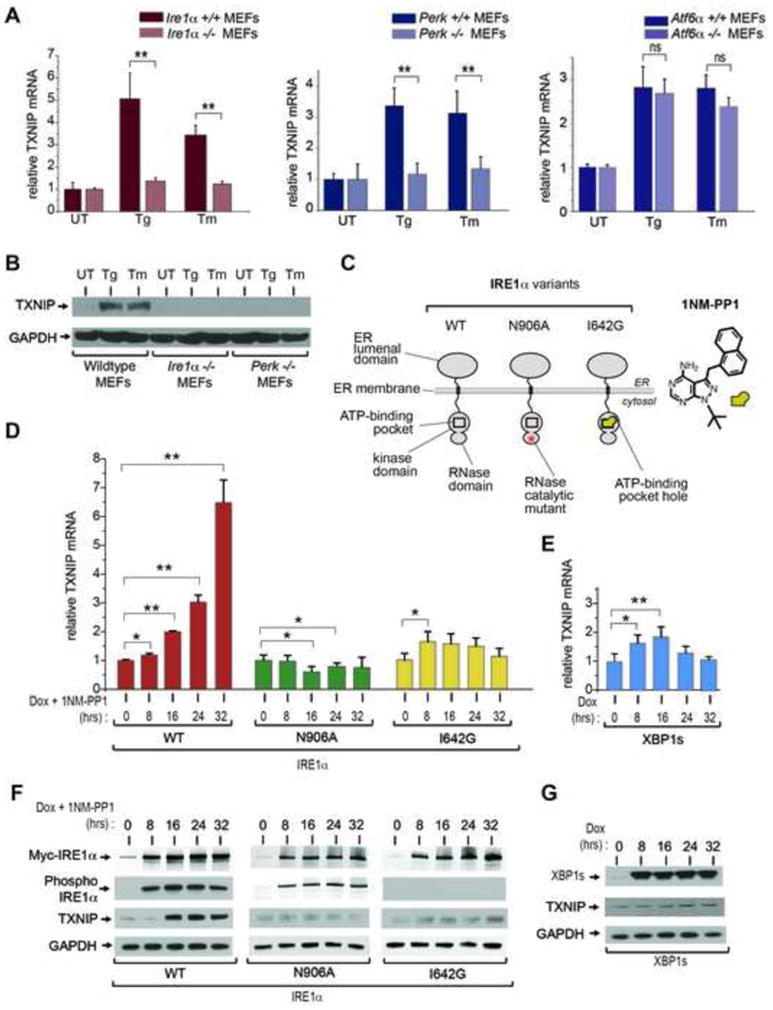
(A) Analysis of TXNIP mRNA expression (normalized to GAPDH) by Q-PCR during ER stress treatment in UPR sensor signaling mutants. Atf6α −/−, Perk−/−, and Ire1α−/− MEFs (and wild-type counterparts) were treated with 1 μM Tg, or 5 μg/ml Tm, for 6 hours. (B) Immunoblot for TXNIP protein from whole cell lysates of wild-type, Ire1α−/− and Perk−/− MEFs untreated or treated with 1 μM Tg or 5 μg/ml Tm for 3 hrs. (C) Schematic representation of IRE1α variants used in this study, and chemical structure of 1NM-PP1. (D) Time course analysis of TXNIP mRNA expression (normalized to GAPDH) by Q-PCR through ER stress-independent forcible activation of IRE1α and mutants, and forced expression of XBP1s, in INS-1 cells with 1μg/ml Dox and 5μM 1NM-PP1. (E) Time course analysis of TXNIP proteins (by immunoblot) following forced activation of IRE1α and mutants, and forced expression of XBP1s, in INS-1 cells untreated or treated with 1μg/ml Dox and 5μM 1NM-PP1. Three independent biological samples were used for Q-PCR experiments. Data are shown as mean ± SD. **p < 0.005, *p < 0.01.
