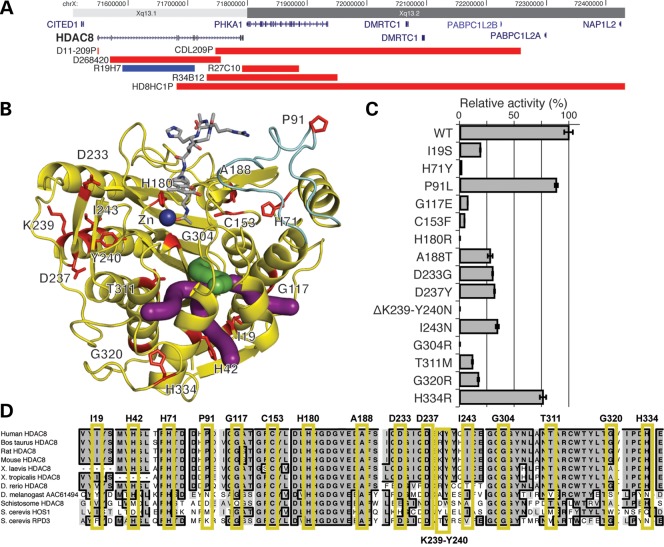
Figure 1.
Mutations in HDAC8. (A) Copy number abnormalities that disrupt the HDAC8 locus. Localization of deletions (red) and duplications (blue) on Xq13 are indicated by chromosome band and position (hg19). Gene locations are indicated in blue with gene names. HDAC8 is indicated in bold. (B) Localization of HDAC8 missense mutations on the crystal structure (PDB accession code 2V5W (27)). Mutated residues are in red and labeled. The acetylated substrate is in gray and the active site zinc ion (Zn) is in dark blue. The light blue portion indicates the L2 substrate-binding loop. Green and purple tubes indicate the catalytically important main and branch exit channels, respectively, for the acetate hydrolysis product (28,29). (C) HDAC8 mutations disrupt deacetylase activity. Bar graphs demonstrate the effect of HDAC8 mutations on deacetylase relative activity relative to the wild-type enzyme activity (specific activities are noted in Supplementary Material, Table S1). All assays were performed in triplicate. Error bars indicate standard deviation. Enzymatic activity for all mutations is significantly less than wild type (P <0.05) using a two-tailed unpaired t-test. Single letter amino acids are used for mutation notation to correlate with residues in (B and C). (D) Conservation of mutations in HDAC8. Alignment of HDAC8 and related deacetylases from 10 species, as indicated on the left. Residues identical to the human residue in a majority of species at each position are in bold, in dark gray and a black border. Those that are similar are shaded in lighter gray, without a border. Each mutated human residue is indicated at the top, with a yellow box surrounding the aligned residues. Note the conservation through yeast for all residues in which a missense mutation causes a complete loss of function.