Abstract
In Brassica napus, seed yield and quality are related to sulfate availability, but the seed metabolic changes in response to sulfate limitation remain largely unknown. To address this question, proteomics and biochemical studies were carried out on mature seeds obtained from plants grown under low sulfate applied at the bolting (LS32), early flowering (LS53), or start of pod filling (LS70) stage. The protein quality of all low-sulfate seeds was reduced and associated with a reduction of S-rich seed storage protein accumulation (as Cruciferin Cru4) and an increase of S-poor seed storage protein (as Cruciferin BnC1). This compensation allowed the protein content to be maintained in LS70 and LS53 seeds but was not sufficient to maintain the protein content in LS32 seeds. The lipid content and quality of LS53 and LS32 seeds were also affected, and these effects were primarily associated with a reduction of C18-derivative accumulation. Proteomics changes related to lipid storage, carbohydrate metabolism, and energy (reduction of caleosins, phosphoglycerate kinase, malate synthase, ATP-synthase β-subunit, and thiazole biosynthetic enzyme THI1 and accumulation of β-glucosidase and citrate synthase) provide insights into processes that may contribute to decreased oil content and altered lipid composition (in favor of long-chain fatty acids in LS53 and LS32 seeds). These data indicate that metabolic changes associated with S limitation responses affect seed storage protein composition and lipid quality. Proteins involved in plant stress response, such as dehydroascorbate reductase and Cu/Zn-superoxide dismutase, were also accumulated in LS53 and LS32 seeds, and this might be a consequence of reduced glutathione content under low S availability. LS32 treatment also resulted in (i) reduced germination vigor, as evidenced by lower germination indexes, (ii) reduced seed germination capacity, related to a lower seed viability, and (iii) a strong decrease of glyoxysomal malate synthase, which is essential for the use of fatty acids during seedling establishment.
As the third main oil crop worldwide (58.5 Mt in 2011), oilseed rape represents a major renewable resource for food (oil, meal) and nonfood uses (green energy, green chemistry). Relative to other crops such as cereals, oilseed rape (Brassica napus L.) requires high amounts of sulfur (S) to sustain its growth and yield (1–3). The reduction of S atmospheric deposits observed over recent decades has forced farmers to add S fertilizer in order to maintain seed yield and quality. A previous study highlighted the necessity of satisfying plant S requirements until the start of pod filling to ensure yield as well as high lipid and protein contents (4). These observations emphasize the importance of a detailed understanding of the impact of S limitation on seed oil and protein quality and of the processes involved.
During Brassica napus seed development, the carbon (C) provided by source organs as sucrose is assimilated through both oxidative phosphate and glycolytic pathways. These pathways provide precursors for fatty acid synthesis in the form of acetyl-CoA, an S-containing metabolite. Glycolysis enables the production of phosphoenolpyruvate (PEP)1 from hexose phosphates formed from sucrose cleavage and is considered as the predominant metabolic pathway for the production of these precursors. During seed development, PEP is principally transported to the plastid, where it is dephosphorylated by pyruvate kinase into pyruvate, which is the substrate responsible for acetyl-CoA formation, used for fatty acid synthesis inside plastids by acetyl-CoA carboxylase and fatty acid synthase (5, 6). In plastids of Brassica napus cells, acetyl-CoA carboxylase, which catalyzes the carboxylation of acetyl-CoA to form malonyl-CoA, needed to sustain de novo fatty acid synthesis (C16:0, C18:0, C18:1), is present both in the prokaryotic form, consisting of a protein complex of four assembled subunits, and in the eukaryotic form, as a single large multifunctional polypeptide (7). In the cytosol, PEP can also produce pyruvate from cytosolic pyruvate kinase or through a system involving PEP carboxylase, malate dehydrogenase (MDH), and chloroplastidial malic enzyme. The PEP carboxylase–MDH–malic enzyme pathway might be important, as PEP carboxylase activity is substantial during Brassica napus seed development, relative to most nonphotosynthetic tissues (8). The cytosolic pyruvate can also be transported to the mitochondria to produce energy through the TCA cycle. Nevertheless, in maturing B. napus embryos, flux through the complete TCA cycle is absent and oxidation of mitochondrial substrate only weakly contributes to ATP production (9). The mitochondrial metabolism is mostly devoted to cytosolic fatty acid elongation, because the citrate formed in the TCA cycle is exported into the cytosol and used for the production of acetyl-CoA by ATP citrate lyase (10). The multifunctional acetyl-CoA carboxylase, also present in the cytosol, provides malonyl-CoA required for fatty acid elongation (C20:0, C20:1, C22:0) and for a variety of reactions including the synthesis of secondary metabolites such as flavonoids and anthocyanins and the malonylation of some amino acids and secondary metabolites (7).
After the extraction of oil from B. napus seeds, the residual protein-rich meal is used for animal feed. Cruciferins are the major form of seed storage protein (SSP) found in Brassica species. These 11–12S globulins are synthesized inside the endoplasmic reticulum during seed development as a precursor form of 50 to 60 kDa, prior to being transported via the Golgi to vacuoles, where they are partially cleaved during a later stage by vacuolar proteases, leading to the formation of acidic α- and basic β-subunits. In dry mature seeds, cruciferins stored in vacuoles are composed of six pairs of acidic and basic associated subunits that interact noncovalently. These subunits are subjected to limited proteolysis at the C-terminal end (11–13), which is repressed by S limitation treatments in Arabidopsis thaliana (14). During germination, SSPs are broken down and used as a source of nitrogen (N), C, and S by the germinating seedling (11). The effects of S limitation on seed protein quality have been studied in Arabidopsis thaliana (14), in which S limitation leads to decreased seed protein content, principally associated with a decrease in S-rich SSP accumulation (At12S3, At2S3). In oilseed rape, the N/S ratio in seed protein increases in S-limited conditions (2). Many attempts have been made to increase the seed methionine content of Brassicaceae species (see Ref. 15 for a review). Transgenic Brassica napus lines carrying a gene encoding the Brazil nut (Bertholletia excelsa) 2S albumin, a methionine-rich storage protein (representing 18.8% of the total amino acids of this protein), and fused with the regulatory region of the phaseolin gene show significant enhancement of total seed methionine accumulation under non-S-limiting conditions (16), improving the nutritional value of Brassica napus seeds. Unfortunately, this Brazil nut 2S sulfur-rich albumin was found to be allergenic (17). Recently, it has been reported that reduced activity in homocysteine methyltransferase 2, a methionine biosynthetic enzyme specifically expressed in vegetative tissues, leads to an increased accumulation of methionine in Arabidopsis thaliana seeds (18). To our knowledge, such an attempt has not yet been made in the context of S deficiency. Moreover, although the effect of S deficiency on the major seed proteins has been investigated in Arabidopsis (14), there has been no report on the effect of S limitation on the seed proteome in Brassica napus L., a major oil crop grown worldwide.
With the knowledge that S limitation leads to perturbations of S, C, and N metabolism (19–22), and considering the importance of such metabolism for lipid and protein synthesis in developing seeds, this study aimed to characterize the effects of S limitation applied at different growth stages on Brassica napus seed quality. In addition, the study reveals the adaptations that may occur during seed maturation in response to S limitation. Their consequences for the maintenance of seed yield, lipid and protein quality, and germination capacity are also discussed.
EXPERIMENTAL PROCEDURES
Experimental Treatments and Tissue Sampling
Brassica napus plants were grown and harvested according to the protocol described by Dubousset et al. (4). Briefly, Brassica napus cv. Capitol seeds were germinated with a thermoperiod of 20 °C (day, 16 h) and 15 °C (night, 8 h) for 36 days on 25% Hoagland nutrient solution. The plants were then subjected to 8 °C (day, 10 h) and 4 °C (night, 14 h) for 46 days for vernalization with the same nutrient solution. Each plant was then transferred to one pot containing perlite and vermiculite (2:1, v/v) and subjected to a thermoperiod of 20 °C (day) and 15 °C (night). Each day, the nutrient solution was supplied automatically in an increasing volume as a function of the growth stage: 90, 120, 150, and 180 ml per plant at the start of the bolting, visible bud, flowering, and seed maturation stage, respectively. Optimal S nutrition (508.7 μm SO42−) was applied in control plants, and mineral S restriction (low-sulfur (LS) treatments) corresponding to 8.7 μm SO42− was applied at GS32 (bolting stage) for LS32 treatment, GS53 (visible bud to early beginning of flowering) for LS53 treatment, or GS70 (start of pod filling) for LS70 treatment, until the end of the growth cycle. At the mature seed stage, the number of mature seeds per plant was determined for four replicates per treatment. Seeds were then used for all the analyses. The yield component data and amounts of total oil, total proteins, and glucosinolates were previously published by Dubousset et al. (4).
Determination of Total S and S-proteic Contents
Freeze-dried samples were ground to a fine powder, weighed, and placed into tin capsules for determination of the total S content using an elemental analyzer (EA3000, EuroVector, Milan, Italy) connected to a continuous-flow isotope ratio mass spectrometer (Isoprime, GV Instruments, Manchester, UK).
For determination of the S-proteic content, soluble proteins were extracted by grinding 30 mg fresh weight of seed in 0.5 ml of citrate Na-phosphate buffer (20 mm citrate and 160 mm Na2HPO4, pH 6.8). The homogenate was centrifuged at 12,000g at 4 °C for 1 h, and the resulting supernatant was used to determine the concentration in soluble proteins by means of protein-dye staining (23), using BSA as a standard. Proteins were then precipitated by the addition of four volumes of 10% TCA in acetone to one volume of extract. After storage at −20 °C overnight, the extract was centrifuged (12,000g, 4 °C, 20 min), and the resulting pellet was washed twice with 1 ml of 80% acetone and centrifuged (16,000g, 4 °C, 3 min). Residual acetone was evaporated under vacuum at 45 °C, and the resulting pellet, resuspended in 0.1 ml of ultrapure water, was placed into a tin capsule. The water was then evaporated under vacuum at 45 °C (SpeedVac Concentrator 5301, Eppendorf, Le Pecq, France), and dry protein extract was analyzed for S content using an elemental analyzer combined with an isotope ratio mass spectrometer as described above for total S content.
Extraction and Quantification of Sulfate Content
Sulfate was extracted from 45 mg dry weight of seed samples ground to a fine powder, incubated twice with 2 ml of 50% ethanol at 40 °C for 1 h, centrifuged (10,000g for 20 min), incubated twice with water at 95 °C for 1 h, and centrifuged again (10,000g for 20 min). The supernatants were pooled and evaporated under vacuum (Concentrator Evaporator RC 10.22, Jouan, Saint-Herblain, France). The dry residue was resuspended in 2 ml of ultrapure water, and the sulfate concentration was determined via HPLC (ICS3000, Dionex Corp., Sunnyvale, CA).
Determination of Free Cysteine and Glutathione Contents via HPLC
Thiols were extracted by grinding 20 mg fresh weight of seed in 0.2 ml of 0.1 m HCl. After centrifugation (20,000g for 10 min), the supernatant was used to measure the content of total free cysteine and total glutathione after reduction with DTT by HPLC using the monobromobimane derivatization method as described by Koprivova et al. (24).
Determination of Oil, Protein, and Glucosinolate Contents via Near-infrared Spectroscopy
As previously described by Dubousset et al. (4), all the seed samples (about 5 g) were scanned using a near-infrared spectrometer (NIRS model 6500, FOSS NIRSystem Inc., Silver Spring, MD) equipped with a transport module in the reflectance mode. The results were predicted from an external calibration established for oil and total glucosinolate (GLS) content (CRAW, Gembloux, Belgium). Three determinations were performed for each sample.
Extraction and Quantification of Oil Content and Lipid Composition
The method for determining seed oil content was based on direct methylation of fatty acids. Briefly, 10 mature dried seeds were ground in a microtube containing three inox balls using a tissue-lyser system (Qiagen, Chatsworth, CA). For each sample, three aliquots of 10 mg each were weighed and transferred into glass tubes with Teflon-lined screw caps containing 1.32 ml of methanol/sulfuric acid/toluene (100/2.5/30, v/v/v) with 400 μg ml−1 of heptadecanoic acid as an internal standard. The mixture was shaken vigorously for 30 s, heated (95 °C for 1 h), and cooled on ice, and fatty acid methyl esters were then extracted into 500 μl of hexane following the addition of 1 ml of water. After vigorous hand shaking (15 s) and centrifugation (650g for 5 min), 10 μl of the upper organic phase was analyzed using gas chromatography. If necessary, extracts were evaporated under nitrogen and dissolved into 50 μl of hexane before gas chromatography analysis. Fatty acid methyl esters were separated on a DB-WAX column (30 ml by 0.25 mm inner diameter, 25-μm film, J&W Scientific Columns, Agilent Technologies Co., Palo Alto, CA) and quantified with a flame ionization detector using the recommendations of the manufacturer (Agilent). To determine the mass of each fatty acid, the peak area was compared with the internal standard peak area.
Two-dimensional Electrophoresis and Image Analysis
Total proteins were extracted from 10 mg of mature seeds, without discarding nonviable seeds, according to a protocol described by Gallardo et al. (13) (n = 6 for control; n = 4 for LS70, LS53, and LS32). After 1 h of incubation at room temperature in thiourea/urea buffer, the extracts were centrifuged twice at 20,000g and 4 °C for 10 min. The protein concentration was then measured according to Bradford's method (23). Isoelectric focalization was performed from 250 μg of proteins as described by Gallardo et al. (25) using gel strips forming an immobilized nonlinear pH 3–10 gradient (Immobiline DryStrip, 24 cm; GE Healthcare, Saclay, France), and proteins were separated in vertical polyacrylamide gels as described by Gallardo et al. (13). Gels were stained with Coomassie Brilliant Blue G-250 (Bio-Rad, Marne la Coquette, France) according to the method of Mathesius et al. (26). Gel image acquisition was performed using an Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE) at 700 nm with a resolution of 169 μm. Image analyses and spot volume quantification were carried out with Progenesis SameSpots v3.0 software (Nonlinear Dynamics, Newcastle upon Tyne, UK) according to the manufacturer's protocol. Molecular weights (Mr) and isoelectric points (pI) were calculated according to the migration of standard proteins (Bio-Rad).
Protein Identification via Electrospray Ionization–Liquid Chromatography-Tandem Mass Spectrometry
Spots of interest were excised, washed several times with water, and dried. Trypsin digestion was performed overnight with a dedicated automated system (MultiPROBE II, PerkinElmer Life Sciences, Courtaboeuf, France). The gel fragments were subsequently incubated twice for 15 min in a 0.1% CH3CN solution to allow extraction of peptides from the gel pieces. Peptide extracts were dried and dissolved in starting buffer for chromatographic elution; the buffer consisted of 3% CH3CN and 0.1% HCOOH. Peptides were enriched and separated using lab-on-a-chip technology (Agilent) and fragmented using an on-line XCT mass spectrometer (Agilent). The software used to generate the peak list was Data Analysis for 6300 series Ion Trap LC/MS (v3.4; Bruker Daltonique, Wissembourg, France). Identification was performed using X!Tandem, developed by the Global Proteome Machine organization, and the NCBI nonredundant protein sequence database filtered for viridiplantae entries (released February 11, 2012; v4; 1,945,867 entries). The parameters used in the X!Tandem search were as follows: enzyme, trypsin; number of missed cleavages, 1; fixed modifications, carbamidomethylation of cysteine; variable modifications, oxidation of methionine; lower window mass tolerance for precursor ions, 1 Da; upper window mass tolerance for precursor ions, +2.5 Da; mass tolerance for fragment ions, 0.5 Da. Identified proteins were filtered using the X!TandemPipeline (http://pappso.inra.fr/bioinfo/xtandempipeline/) with the following criteria: (i) two or more different peptides with an E value less than 0.05 and (ii) a protein log E value (the log of the product of unique peptide E values) less than 4. The results of proteomic analysis and protein identification were incorporated into the PROTICdb database (27), a Web-based application that allows storage and querying of information related to proteomic analysis (http://moulon.inra.fr/protic/sms). These results are also available in a synthetic form (supplemental Table S3). The identified proteins for spot 233 were filtered on the basis of only one peptide and subsequently a protein log E value less than 1, because of the small size of the identified protein (79 amino acids; see supplemental Table S4).
Germination Tests
Fifty seeds per biological repetition (n = 4) were sown on Whatman filter paper soaked with 10 ml of sterile water within Petri dishes (12 × 12 cm). The Petri dishes were then closed and placed in darkness inside a growth chamber at 20 °C and 70% relative humidity. Seeds that showed a completed radicle protrusion through the seed coat were counted as germinated. The observation was conducted at regular time steps, and the final germination rate was determined at 58 h. The time to reach 5%, 10%, and 50% of the final germination rate (T′5, T′10, and T′50) was calculated using the Gompertz functions (28). The coefficient of the rate of germination was calculated according to Bewley and Black (11) using the following equation:
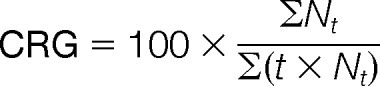 |
where Nt is the proportion of germinated seeds (cumulative germination rate) observed at time t.
Tetrazolium Assay
The viability of seeds was estimated using a tetrazolium assay adapted from the procedure described by Wharton (29). After 5 h of incubation in water at 30 °C in darkness, seeds were scarified and incubated in a 1% (w/v) aqueous solution of 2,3,5-triphenyltetrazolium chloride (Sigma Aldrich, Saint Quentin Fallavier, France) at 30 °C in darkness for 18 h. For treatment (control, LS70, LS53, and LS32), the tetrazolium test was performed on 20 seeds (5 seeds of each biological replication, n = 4). If a seed is viable, tetrazolium salts are metabolically reduced to highly red-colored end products called formazans by NADH-dependent reductases of the endoplasmic reticulum. Pink staining or a white zone on the radicle or on cotyledons indicates reduced seed viability (30).
Statistics
The variability of the results is expressed by the average values for all biological replicates (n = 3, 4, or 6) ± the standard error (S.E.). The effects of LS70, LS53, and LS32 treatments relative to the control conditions were subjected to statistical analysis using Microsoft® Excel 2008/XLSTAT©-Pro (Version 7.2, Addinsoft, Inc., Brooklyn, NY). With a statistical significance postulated at p < 0.05, the Mann–Whitney test was done to compare data obtained for LS and control seeds. Protein spots specifically induced and repressed in LS70, LS53, and LS32 seeds were subsequently analyzed via electrospray ionization–liquid chromatography–tandem mass spectrometry.
RESULTS
Effect of S Limitation on Yield Components, Quality, and S Status in Mature Seeds
Seed yield was significantly reduced by S limitation applied at the bolting stage (LS32 treatment) but not by S limitation applied at later stages (LS53 and LS70 conditions) (Table I). The number of pods per plant was not affected, but the thousand-seed weight was reduced by 33% in LS32 treatment relative to the control (Table I). Similarly, LS32 treatment led to an altered seed morphology (Fig. 1A). Furthermore, the seed water content at harvest was below the threshold of 10% for both control and LS conditions, with only a slightly lower water content for LS32 seeds (7.5% ± 0.1) than for control mature seeds (6.0% ± 0.1; Table II).
Table I. Yield components and contents of oil, proteins, and glucosinolates in mature seeds produced by plants grown under control, LS70, LS53, and LS32 conditions (4).
| Total seed DW produced per plant (g) | Maximal pod length (cm) | Number of pods per plant | Thousand-seed weight (mg) | Oil content in mature seeds (% DW) | Protein content in mature seeds (% DW) | Glucosinolate content in mature seeds (μmol g−1 DW) | |
|---|---|---|---|---|---|---|---|
| Control | 11.6 ± 0.61 | 8.3 ± 0.1 | 178 ± 4 | 4694 ± 39.50 | 45 ± 0.5 | 23 ± 0.3 | 14 ± 0.4 |
| LS70 | 11.7 ± 0.49 | 7.9 ± 0.1 | 187 ± 7 | 4583 ± 53.02 | 45 ± 0.2 | 23 ± 0.2 | 8.3 ± 0.5 |
| LS53 | 11.6 ± 0.81 | 8.4 ± 0.1 | 165 ± 9 | 4928 ± 140.1 | 43 ± 0.3 | 22 ± 0.2 | 2.5 ± 0.4 |
| LS32 | 6.30 ± 0.66 | 7.5 ± 0.8 | 162 ± 6 | 3124 ± 293.9 | 32 ± 1.3 | 21 ± 0.3 | 4.2 ± 0.9 |
The values are shown as mean ± S.E. (n = 6 for control; n = 4 for LS70, LS53, and LS32). Significant differences from the control value are indicated in bold (p < 0.05). LS70, LS53, and LS32 treatments respectively correspond to a sulfur limitation applied when the first petals fell (GS70), when the main inflorescence emerged (GS53), and at the beginning of the bolting stage (GS32).
Fig. 1.

Effects of control, LS70, LS53, and LS32 treatments on seed morphology and viability. A, morphology of mature seeds harvested from plants grown under control, LS70, LS53, and LS32 conditions. LS70, LS53, and LS32 treatments corresponded to sulfur limitation applied when the first petals fell (GS70), when the main inflorescence emerged (GS53), and at the beginning of the bolting stage (GS32). B, Brassica napus embryos isolated from control, LS70, LS53, and LS32 seeds and subjected to the tetrazolium assay. White arrows indicate unviable seeds in control, LS70, and LS53 seeds. For details about the method used, see the “Experimental Procedures” section.
Table II. Water content at harvest and germination indexes of Brassica napus seeds produced by plants grown under control, LS70, LS53, and LS32 conditions.
| Water content (%) | Final germination rate (%)a | T′5 (h)b | T′10 (h)b | T′50 (h)b | CRGc | |
|---|---|---|---|---|---|---|
| Control | 6 ± 0.1 | 93.3 ± 2.8 | 21.2 ± 0.3 | 22.6 ± 0.2 | 29.4 ± 1.0 | 2.37 ± 0.01 |
| LS70 | 5.9 ± 0.1 | 92.3 ± 2.3 | 23.2 ± 0.3 | 24.2 ± 0.3 | 28.9 ± 0.9 | 2.37 ± 0.02 |
| LS53 | 6 ± 0.2 | 91.5 ± 2.1 | 23.1 ± 0.3 | 24.5 ± 0.3 | 30.8 ± 1.0 | 2.32 ± 0.01 |
| LS32 | 7.5 ± 0.1 | 40.3 ± 3.5 | 24.1 ± 0.3 | 26.3 ± 0.3 | 36.5 ± 1.2 | 2.22 ± 0.02 |
The values are shown as mean ± S.E. (n = 4). Significant differences from the control value are indicated in bold (p < 0.05).
a Determined at 58 h.
b Time required in order to achieve 5% (T′5), 10% (T′10), or 50% (T′50) germination, calculated from the Gompertz equation (28).
c Coefficient of the rate of germination, as calculated by Bewley and Black (11).
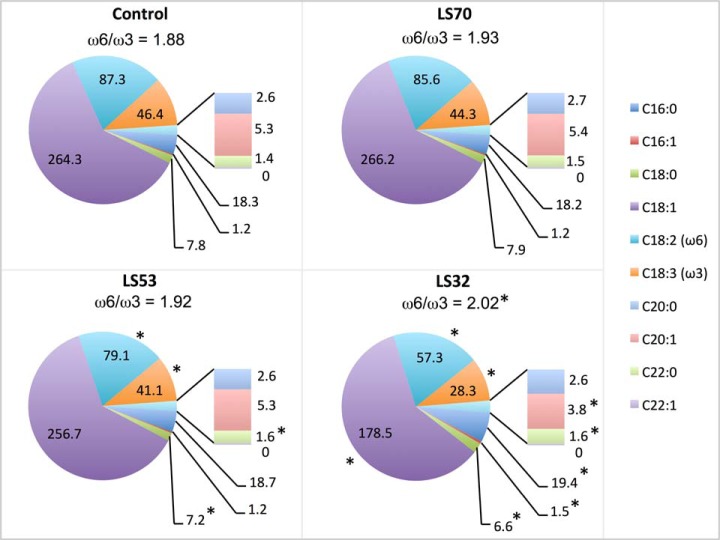
The total oil content was not affected by LS70 treatments but was reduced in LS53 and LS32 seeds (Table I). Whatever the S fertilization treatment, the main fatty acid was oleic acid (C18:1) (Fig. 2, supplemental Table S1). The LS70 treatment had no effect on fatty acid composition in mature seeds. In contrast, LS32 treatment led to a significant reduction of all C18 derivatives. For instance, the quantity of C18:1 in LS32 seeds decreased by 32% relative to the control. In contrast, quantities of palmitoleic acid (C16:1) and behenic acid (C22:0) increased noticeably in response to LS32 treatment. Whereas the behenic acid concentration slightly increased in response to LS53 treatment, the concentrations of stearic (C18:0), linoleic (C18:2), and linolenic (C18:3) acids were significantly reduced relative to the control, but to a lesser extent than in LS32 conditions (Fig. 2, supplemental Table S1). The ω6/ω3 ratio was significantly higher in LS32 seeds than in the control (Fig. 2). The protein content was also significantly lower only in LS32 seeds relative to the control (Table I).
Fig. 2.
Fatty acid composition of mature seeds harvested from plants grown under control, LS70, LS53, and LS32 conditions. The values are shown as the mean in mg g−1 DW (n = 6 for control; n = 4 for LS70, LS53, and LS32). The ω6/ω3 ratios are indicated above the pie charts. *Significant differences from the control value (p < 0.05). Means ± S.E. are indicated in supplemental Table S1.
As expected, the three LS treatments led to significant reductions of S content in mature seeds (as much as 67% in LS32 seeds relative to the control; Table III), together with reductions of S-sulfate (93% in LS32 seeds; Table III) and glucosinolate contents (70% in LS32 seeds; Table I). As for the glutathione content, the S-sulfate/S-total ratio was significantly reduced in LS53 and LS32 seeds relative to the control, but it was similar to the control in LS70 seeds. Interestingly, the free cysteine content was significantly greater in LS53 and LS32 than in control seeds (Table III).
Table III. Sulfur status of mature seeds harvested from plants grown under control, LS70, LS53, and LS32 conditions.
| S content (μmol g−1 DW) | S-sulfate content (μmol g−1 DW) | S-glutathione content (μmol g−1 DW) | Free S-cysteine content (nmol g−1 DW) | S-sulfate/S (%) | |
|---|---|---|---|---|---|
| Control | 121.74 ± 1.81 | 18.38 ± 0.67 | 2.46 ± 0.21 | 2.31 ± 0.13 | 15.12 ± 0.56 |
| LS70 | 88.01 ± 2.84 | 11.58 ± 1.04 | 2.16 ± 0.24 | 2.42 ± 0.50 | 13.13 ± 1.01 |
| LS53 | 62.31 ± 4.59 | 4.63 ± 0.82 | 1.59 ± 0.32 | 3.49 ± 0.11 | 7.29 ± 0.92 |
| LS32 | 39.46 ± 2.75 | 1.28 ± 0.27 | 0.90 ± 0.10 | 4.38 ± 0.38 | 3.21 ± 0.62 |
The values are shown as mean ± S.E. (for S and S-sulfate contents: n = 6 for control and n = 4 for LS70, LS53, and LS32; for S-glutathione and free S-cysteine content, n = 3). Significant differences from the control values are indicated in bold (p < 0.05).
The germination characteristics of control and LS seeds (Table II) were studied. Relative to control seeds, the start of germination was delayed for all LS seeds, and the times required to achieve 5% and 10% germination (T′5 and T′10) were longer than in the control. LS32 seeds also showed a lower coefficient of the rate of germination, which is consistent with the longer time needed for these seeds to reach 50% of the final germination rate (T′50 of 36.5 h) than for control seeds (29.4 h). The final germination rate was not affected in LS70 and LS53 seeds, but it was significantly reduced in LS32 seeds (40.3%) relative to control seeds (93.3%). All seed samples were subjected to a tetrazolium test (Fig. 1B). Interestingly, whereas the viability of LS70 and LS53 seeds was not affected, that of LS32 seeds was strongly reduced, with only 33% of seeds red-colored and thus highly viable.
Modifications in the Seed Proteome in Response to S Limitation
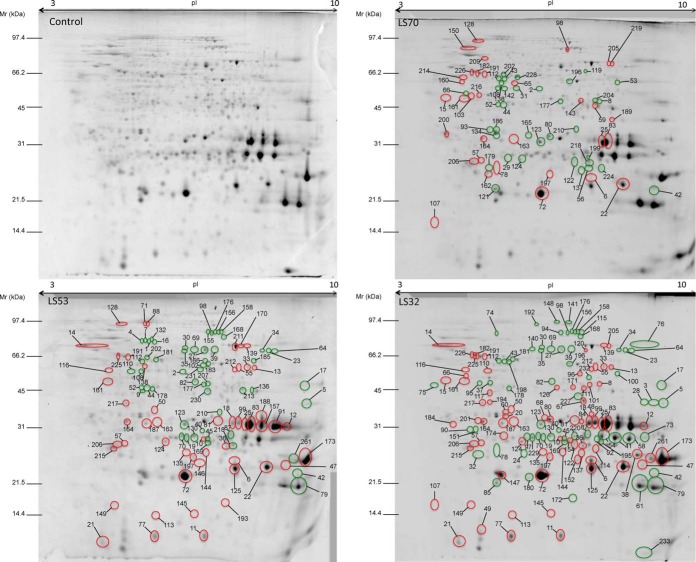
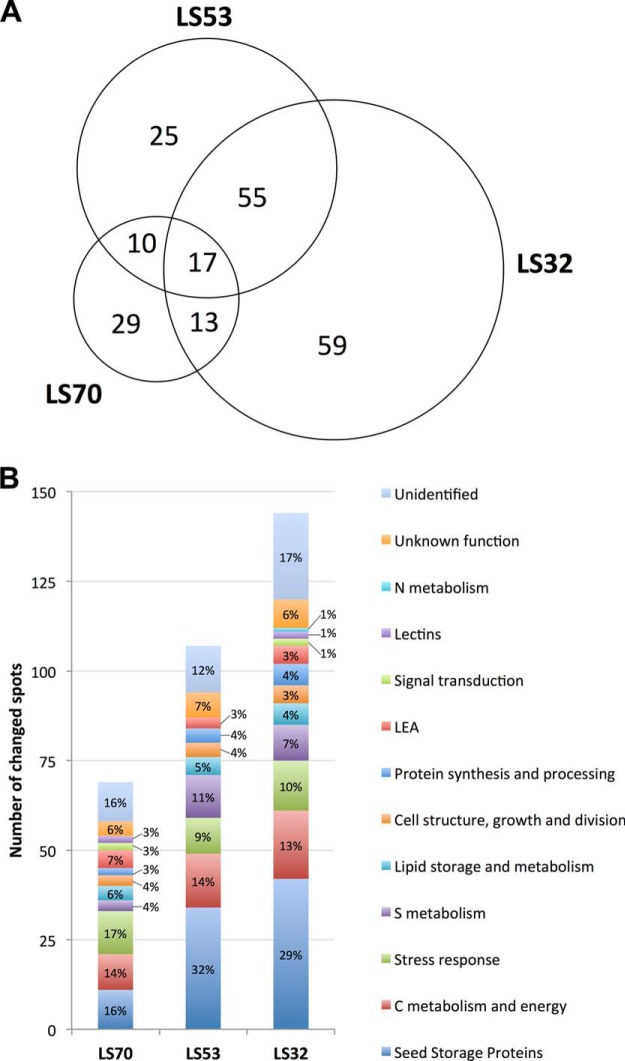
The proteomic analysis resulted in 495 protein spots detected in mature seeds. Among them, 208 protein spots were differentially expressed in mature seeds of S-limited plants relative to the control (Fig. 3). The three LS treatments applied at different growth stages led to different proteomic responses at the mature seed level; only a small proportion of the modulated proteins (17 spots, or 8.17%) were shared by all treatments (Fig. 4A). The number of proteins presenting a significant change in abundance increased with the precocity of S limitation: relative to the control, there were 69 modulated spots for LS70 seeds, versus 144 modulated protein spots for LS32 seeds (Figs. 4A and 4B). Liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) enabled the identification of 83% of the 208 modulated protein spots. All major proteins (i.e. SSPs) detected in the two-dimensional electrophoresis gels were identified in this study (Fig. 3), providing a global view of the SSP component of oilseed rape. Except for the proteins that were not identified or associated with a functional group (supplemental Table S2), these spots could be classified into 11 groups, corresponding to their functional role (Fig. 4B, Tables IV–VII). Among the 11 functional groups, that of SSPs represented 16% of the total protein changes observed in LS70 seeds, versus 32% and 29% in LS53 and LS32 seeds, respectively (Fig. 4B). Similar variation was observed for proteins involved in S metabolism, which represented 4% of the total protein changes in LS70 seeds versus 11% in LS53 seeds and 7% in LS32 seeds (Fig. 4B). These results indicate that SSPs and S metabolism are mainly and gradually affected by the severity of S limitation.
Fig. 3.
Two-dimensional electrophoresis gels of total proteins from control, LS70, LS53, and LS32 mature Brassica napus seeds. For each treatment, the induced and repressed protein spots are circled in red and green, respectively, on the corresponding gel image. These spots were identified by LC-MS/MS and are listed in Tables IV through VII. Unidentified spots or spots that were not associated with a functional group are listed in supplemental Table S2. Mr, molecular weight; pI, isoelectric point.
Fig. 4.
Effects of control, LS70, LS53, and LS32 treatments on mature seed proteome. A, distribution of the 208 proteins presenting a significant variation of abundance in LS70, LS53, and LS32 seeds relative to the control. B, number and distribution (%) in the functional groups of proteins changed in LS70, LS53, and LS32 seeds relative to the control.
Table IV. Significantly changed proteins in LS70, LS53, and LS32 mature seeds identified by mass spectrometry as seed storage proteins, proteins involved in protein synthesis and processing, or late embryogenesis abundant proteins.
| Spot number | Protein name | LS70 | LS53 | LS32 | Species | NCBI accession number | log (E value) | CV (%) | Exp. pI | Exp. Mr (kDa) | Theo. pI | Theo. Mr (kDa) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Seed storage proteins | ||||||||||||
| 4 | Convicilin | −3.3 | −5.5** | −2.0 | Ps | 7339551 | −20.0 | 16 | 5.4 | 77.7 | 5.5 | 72.06 |
| 5 | Cruciferin/CRU1 | 1.3 | −2.4** | −4.4** | Bn | 461840 | −10.2 | 4 | 8.8 | 42.6 | 7.64 | 56.50 |
| 6 | Cruciferin/BnC1 | 1.6* | 2.2** | 5.5** | Bn | 1345840 | −45.6 | 21 | 6.6 | 24.2 | 6.84 | 53.82 |
| 7 | Convicilin | −2.8 | −4.6** | −1.9 | Ps | 227928 | −18.4 | 12 | 5.4 | 78.1 | 5.49 | 71.41 |
| 9 | Legumin | −4.2 | −4.6* | −1.5 | Ps | 126161 | −11.4 | 12 | 5.4 | 48.2 | 6.21 | 59.27 |
| 10 | Cruciferin/CRU1 | 1.5 | 2.2** | 3.9** | Bn | 461840 | −10.6 | 9 | 6.1 | 30.4 | 7.64 | 56.50 |
| 11 | Cruciferin/BnC1 | 1.0 | 2.5** | 3.6** | Bn | 1345840 | −10.4 | 6 | 6.1 | 12.2 | 6.84 | 53.82 |
| 12 | Cruciferin/BnC1 | 1.9 | 3.2** | 2.0* | Bn | 1345840 | −23.5 | 20 | 8.0 | 34.4 | 6.84 | 53.82 |
| 13 | Cruciferin/BnC1 homolog | −1.2 | 1.5* | 2.6** | Bn | 294979712 | −35.5 | 29 | 6.9 | 60.0 | 6.6 | 51.32 |
| 18 | Cruciferin/BnC1 | 1.4 | 2.1** | 3.1** | Bn | 1345840 | −23.7 | 19 | 6.5 | 35.3 | 6.84 | 53.82 |
| 21 | Cruciferin/CRU1 | 1.2 | 2.6** | 3.0** | Bn | 461840 | −17.7 | 14 | 5.0 | 11.4 | 7.64 | 56.50 |
| 22 | Cruciferin/BnC1 | 1.7* | 2.6** | 3.0** | Bn | 1345840 | −50.0 | 25 | 7.3 | 23.2 | 6.84 | 53.82 |
| 25 | Cruciferin/BnC1 | 1.7* | 2.5** | 3.0** | Bn | 1345840 | −40.2 | 20 | 6.8 | 33.9 | 6.84 | 53.82 |
| 32 | Cruciferin/CRU1 | 1.1 | −1.3 | −2.6** | Bn | 461840 | −36.6 | 30 | 5.2 | 26.2 | 7.64 | 56.50 |
| 33 | Cruciferin/BnC1 | 1.2 | 1.3* | 2.8** | Bn | 1345840 | −45.1 | 32 | 6.8 | 60.0 | 6.84 | 53.82 |
| 36 | Cruciferin | 1.5 | 2.0* | 2.7** | Bn | 12751302 | −7.6 | 6 | 6.4 | 28.5 | 8.13 | 54.38 |
| 38 | Cruciferin/CRU1 | 1.2 | 1.3 | 2.7** | Bn | 461840 | −35.2 | 22 | 7.8 | 26.1 | 7.64 | 56.50 |
| 41 | Cruciferin/CRU4 homolog | 1.1 | −1.1 | −2.4** | Al | 297843196 | −60.5 | 18 | 7.0 | 30.1 | 7.09 | 50.65 |
| 42 | Cruciferin/CRU4 | −2.1* | −2.6** | −2.4** | Bn | 17807 | −32.9 | 59 | 8.5 | 21.7 | 8.98 | 20.36 |
| 45 | Cruciferin/CRU4 | 1.0 | −1.2* | −2.4** | Bn | 461841 | −12.6 | 10 | 6.1 | 30.6 | 7.7 | 51.38 |
| 47 | Cruciferin/CRU1 | 1.3 | 1.7* | 2.5** | Bn | 461840 | −9.8 | 10 | 8.1 | 23.8 | 7.64 | 56.50 |
| 48 | Cruciferin/CRU1 | −1.2 | −1.1 | 2.1* | Bn | 461840 | −32.7 | 17 | 6.6 | 38.5 | 7.64 | 56.50 |
| 49 | Cruciferin/CRU1 | 1.0 | 2.0 | 2.4* | Bn | 461840 | −24.7 | 12 | 5.2 | 12.6 | 7.64 | 56.50 |
| 52 | Legumin | −1.8** | −2.4** | −2.0 | Ps | 294979728 | −6.0 | 10 | 5.4 | 49.6 | 6.06 | 56.59 |
| 54 | Cruciferin/CRU4 | 1.1 | −1.3 | −2.2** | Bn | 17805 | −60.1 | 30 | 6.8 | 30.7 | 8.84 | 45.54 |
| 55 | Cruciferin/BnC1 | 1.4 | 1.5** | 2.4** | Bn | 1345840 | −19.1 | 19 | 6.7 | 60.2 | 6.84 | 53.82 |
| 58 | Cruciferin/CRU4 | −1.2 | −1.2 | −2.3** | Bn | 17805 | −71.0 | 31 | 7.5 | 30.3 | 8.84 | 45.54 |
| 61 | Cruciferin/CRU4 | 1.1 | −1.1 | −2.0** | Bn | 17807 | −44.4 | 60 | 7.9 | 19.4 | 8.98 | 20.36 |
| 72 | Cruciferin/BnC1 | 1.7* | 2.2** | 2.1** | Bn | 1345840 | −45.5 | 24 | 5.8 | 21.2 | 6.84 | 53.82 |
| 73 | Cruciferin/CRU4 | 1.2 | −1.0 | −1.8** | Bn | 461841 | −39.0 | 22 | 8.1 | 30.9 | 7.7 | 51.38 |
| 77 | Cruciferin/BnC1 | −1.1 | 1.7* | 1.9** | Bn | 1345840 | −16.1 | 10 | 5.5 | 12.1 | 6.84 | 53.82 |
| 79 | Cruciferin/CRU4 | 1.2 | −1.3* | −1.8** | Bn | 461841 | −39.9 | 20 | 8.4 | 19.1 | 7.7 | 51.38 |
| 83 | Cruciferin/BnC1 | 1.5* | 1.9** | 2.1** | Bn | 1345840 | −35.8 | 21 | 6.8 | 34.3 | 6.84 | 53.82 |
| 85 | Cruciferin | −1.0 | −1.2 | −2.0** | At | 15219584 | −12.9 | 10 | 5.4 | 20.0 | 5.47 | 49.67 |
| 87 | Cruciferin/CRU4 homologue | −1.2 | −1.5* | −2.0** | Al | 297843196 | −55.6 | 17 | 6.5 | 31.6 | 7.09 | 50.65 |
| 91 | Cruciferin/BnC1 | 1.4 | 2.0** | 1.4 | Bn | 1345840 | −25.3 | 22 | 7.6 | 33.7 | 6.84 | 53.82 |
| 92 | Cruciferin/CRU4 homologue | −1.1 | −1.1 | −2.0** | Al | 297843196 | −11.3 | 14 | 6.7 | 30.1 | 7.09 | 50.65 |
| 99 | Cruciferin/BnC1 | 1.2 | 1.5* | 1.9** | Bn | 1345840 | −19.6 | 14 | 6.6 | 35.4 | 6.84 | 53.82 |
| 121 | Cruciferin/BnC1 | −1.5* | 1.2 | 1.1 | Bn | 1345840 | −38.9 | 17 | 5.4 | 22.0 | 6.84 | 53.82 |
| 125 | Cruciferin/BnC1 | 1.3 | 1.6* | 1.8* | Bn | 1345840 | −111.1 | 30 | 6.6 | 22.9 | 6.84 | 53.82 |
| 132 | Convicilin | −1.7 | −1.7** | −1.6 | Ps | 7339551 | −27.8 | 18 | 5.5 | 78.4 | 5.5 | 72.06 |
| 147 | Cruciferin/BnC1 | −1.2 | 1.2 | 1.3* | Bn | 1345840 | −41.6 | 19 | 5.4 | 21.7 | 6.84 | 53.82 |
| 157 | Cruciferin | 1.2 | 1.6* | 1.3 | Bn | 12751302 | −40.3 | 16 | 7.1 | 34.8 | 8.13 | 54.38 |
| 173 | Cruciferin/CRU1 | 1.4 | 1.3* | 1.5** | Bn | 461840 | −81.6 | 32 | 8.7 | 24.4 | 7.64 | 56.50 |
| 174 | Cruciferin/CRU1 | −1.3 | 1.1 | 1.2** | Bn | 461840 | −17.6 | 13 | 5.5 | 31.5 | 7.64 | 56.50 |
| 180 | Cruciferin/BnC1 | −1.0 | 1.1 | −1.4** | Bn | 1345840 | −19.6 | 17 | 5.7 | 21.1 | 6.84 | 53.82 |
| 188 | Cruciferin/CRU1 | 1.1 | 1.4* | 1.5 | Bn | 461840 | −75.5 | 21 | 7.1 | 36.9 | 7.64 | 56.50 |
| 195 | Cruciferin/CRU4 | −1.0 | 1.0 | −1.4** | Bn | 461841 | −60.8 | 24 | 6.7 | 29.7 | 7.7 | 51.38 |
| 197 | Cupin | 1.5* | 1.4* | 1.4** | At | 15226403 | −10.4 | 9 | 5.9 | 25.5 | 5.83 | 55.71 |
| 199 | Cruciferin/CRU4 | −1.4** | −1.2 | −1.0 | Bn | 461841 | −22.0 | 16 | 6.5 | 30.1 | 7.7 | 51.38 |
| 210 | Cruciferin/CRU1 | −1.3* | −1.4* | −1.1 | Bn | 461840 | −10.8 | 8 | 6.3 | 39.4 | 7.64 | 56.50 |
| 233 | Napin | −2.1 | −3.1 | −4.7* | Bn | 243209 | −1.6 | 15 | 8.1 | 10.4 | 8.31 | 8.91 |
| 261 | Cruciferin/CRU1 | 1.4 | 2.0 | 1.5* | Bn | 461840 | −55.1 | 30 | 8.4 | 25.2 | 7.64 | 56.50 |
| Protein synthesis and processing | ||||||||||||
| 100 | Elongation factor 1 α | 1.2 | −1.4 | −1.6* | Bn | 241740165 | −6.3 | 7 | 7.1 | 56.9 | 9.19 | 49.48 |
| 108 | Translational initiation factor | −1.4** | −1.3* | 1.3 | At | 18400210 | −26.7 | 19 | 5.4 | 58.0 | 5.47 | 46.70 |
| 110 | Translational initiation factor | −1.2 | −1.4** | 1.3** | At | 15221761 | −17.7 | 15 | 5.3 | 58.0 | 5.45 | 46.76 |
| 140 | Chaperonin | −1.1 | −1.1 | −1.7** | At | 15229866 | −16.6 | 16 | 5.8 | 71.7 | 6.03 | 59.78 |
| 142 | Translational initiation factor | −1.4** | −1.1 | 1.2 | At | 18400210 | −30.3 | 22 | 5.5 | 58.1 | 5.47 | 46.70 |
| 168 | Glycosyl hydrolase family 38 protein | 1.0 | −1.2* | −1.5** | Al | 297807421 | −35.5 | 12 | 6.4 | 80.6 | 6.21 | 116.02 |
| 194 | 60S acidic ribosomal protein | 1.2 | 1.1 | 1.5** | At | 15229706 | −18.1 | 25 | 5.3 | 42.9 | 5.05 | 34.39 |
| 217 | 60S acidic ribosomal protein | 1.2 | 1.2* | 1.3** | At | 15229706 | −21.1 | 37 | 5.2 | 42.8 | 5.05 | 34.39 |
| Late embryogenesis abundant proteins | ||||||||||||
| 98 | Late embryogenesis-abundant protein | 1.3* | −1.1* | −1.5** | At | 15227965 | −11.5 | 2 | 6.2 | 83.3 | 5.78 | 67.20 |
| 137 | Late embryogenesis-abundant protein 1b | −1.4* | −1.0 | 1.2* | Bn | 79150665 | −13.6 | 13 | 6.4 | 26.6 | 6.02 | 16.67 |
| 182 | Late embryogenesis-abundant protein | 1.2* | 1.1 | 1.5** | At | 15231736 | −10.6 | 7 | 5.2 | 67.5 | 5.29 | 52.08 |
| 191 | Late embryogenesis-abundant protein | 1.3* | 1.3* | 1.5** | At | 15231736 | −10.1 | 9 | 5.2 | 66.6 | 5.29 | 52.08 |
| 200 | Seed maturation protein | 1.3* | 1.0 | −1.1 | At | 15228768 | −9.3 | 9 | 4.7 | 37.6 | 4.73 | 26.74 |
| 225 | Late embryogenesis-abundant protein | 1.2 | 1.2** | 1.3** | At | 15231736 | −16.0 | 10 | 5.1 | 67.2 | 5.29 | 52.08 |
Significant analysis of variance was followed by a Mann–Whitney test (p < 0.05) carried out on the normalized spot volumes (n = 6 for control; n = 4 for LS70, LS53, and LS32). Values indicate fold changes in protein abundance in LS seeds relative to control seeds on a linear scale. Significant differences from the control were at *p < 0.05 or **p < 0.01. The assigned best-matched protein is listed with the organism in which it was identified and its GenBank protein accession number. The log (E value), the percentage of sequence coverage (CV), and experimental (Exp.) and theoretical (Theo.) pI/Mr values obtained are also indicated.
Table V. Significantly changed proteins in LS70, LS53, and LS32 mature seeds identified by mass spectrometry as involved in sulfur metabolism; nitrogen metabolism; cell structure, growth, and division; and signal transduction.
| Spot number | Protein name | LS70 | LS53 | LS32 | Species | NCBI accession number | log (E value) | CV (%) | Exp. pI | Exp. Mr (kDa) | Theo. pI | Theo. Mr (kDa) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S metabolism | ||||||||||||
| 1 | Myrosinase | −3.3 | −7.5** | −1.9 | Bn | 5459292 | −12.8 | 11 | 5.5 | 77.7 | 6.32 | 60.42 |
| 17 | Myrosinase-associated protein | −1.3 | −2.1** | −3.1** | Bn | 1216389 | −50.9 | 38 | 8.7 | 51.2 | 8.47 | 41.82 |
| 27 | Myrosinase 1 | −1.1 | −1.5** | −3.0** | Bn | 310781304 | −43.1 | 24 | 6.1 | 70.6 | 6.62 | 62.74 |
| 30 | Myrosinase 2 | −1.2 | −1.6** | −2.9** | Bn | 310781306 | −25.5 | 18 | 5.9 | 71.8 | 6.45 | 62.72 |
| 31 | Myrosinase-binding protein | −2.4** | 1.2 | 1.2 | Bn | 1655824 | −31.4 | 9 | 5.6 | 57.4 | 5.48 | 99.46 |
| 35 | Myrosinase | −1.1 | −1.5** | −2.7** | Bn | 127733 | −36.0 | 23 | 5.8 | 71.4 | 6.62 | 62.74 |
| 40 | Thiazole biosynthetic enzyme | −1.2 | −1.5* | −2.6** | At | 15239735 | −9.5 | 18 | 6.0 | 33.2 | 5.82 | 36.66 |
| 65 | Myrosinase-binding protein | 2.1** | 1.1 | −1.1 | Bn | 1655826 | −25.3 | 10 | 5.6 | 59.7 | 5.77 | 67.30 |
| 69 | Myrosinase | −1.3 | −1.6** | −2.2** | Br | 56130949 | −30.2 | 19 | 5.9 | 71.4 | 6.65 | 62.97 |
| 71 | Myrosinase-binding protein | 1.2 | 1.6* | −1.4 | Bn | 1655824 | −60.8 | 27 | 5.4 | 91.5 | 5.48 | 99.46 |
| 76 | Myrosinase | 1.1 | −1.3 | −2.0** | Bn | 127733 | −8.2 | 6 | 8.0 | 74.8 | 6.62 | 62.74 |
| 88 | Myrosinase-binding protein | 1.2 | 1.5* | −1.3 | Bn | 1655824 | −35.0 | 15 | 5.5 | 91.5 | 5.48 | 99.46 |
| 138 | Myrosinase-binding protein | −1.5** | 1.2* | 1.0 | Bn | 1655824 | −29.3 | 15 | 5.4 | 61.3 | 5.48 | 99.46 |
| 158 | Cobalamin-independent methionine synthase | 1.2 | −1.2* | −1.4** | At | 15238686 | −36.3 | 19 | 6.4 | 84.7 | 6.09 | 84.36 |
| 176 | Cobalamin-independent methionine synthase | 1.2 | −1.3** | −1.2** | Al | 297807807 | −29.1 | 19 | 6.3 | 84.5 | 6.12 | 84.35 |
| 227 | Epithiospecifier protein | −1.1 | −1.1 | −1.3** | Br | 211905345 | −34.1 | 36 | 6.4 | 45.8 | 5.95 | 37.91 |
| N metabolism | ||||||||||||
| 37 | Glycine cleavage T-protein family | 1.3 | −1.1 | −2.0** | At | 79470337 | −12.1 | 14 | 5.3 | 51.9 | 6.3 | 43.53 |
| Cell structure, growth, and division | ||||||||||||
| 14 | β-glucosidase | 1.2 | 1.9* | 3.2** | Bn | 757740 | −8.5 | 8 | 4.8 | 73.4 | 6.21 | 58.51 |
| 95 | Actin (actin 7) | 1.2 | −1.2 | −1.7** | Bn | 4139264 | −15.4 | 24 | 5.2 | 51.7 | 5.29 | 41.69 |
| 117 | Annexin 1 | 1.0 | 1.1 | 1.8** | Bn | 300433289 | −17.5 | 24 | 5.3 | 47.2 | 5.34 | 36.16 |
| 152 | Actin (actin 11) | 1.1 | 1.2 | 1.6** | Cc | 111610552 | −7.2 | 16 | 6.4 | 24.8 | 4.78 | 23.36 |
| 160 | β-tubulin | 1.4** | −1.1 | 1.1 | At | 18424620 | −15.5 | 16 | 5.0 | 62.2 | 4.7 | 50.73 |
| 161 | β-tubulin | 1.4** | 1.6** | 1.5** | Bn | 8050828 | −6.6 | 10 | 5.0 | 52.3 | 5.69 | 36.10 |
| 170 | β-glucosidase | 1.3 | 1.5** | 1.3 | Bn | 757740 | −37.3 | 29 | 6.7 | 73.7 | 6.21 | 58.51 |
| 211 | β-glucosidase | 1.1 | 1.4** | 1.1 | Bn | 757740 | −42.1 | 30 | 6.6 | 72.3 | 6.21 | 58.51 |
| 214 | β-tubulin | 1.2* | 1.1 | −1.1 | At | 18424620 | −6.9 | 5 | 5.0 | 64.0 | 4.7 | 50.73 |
| Signal transduction | ||||||||||||
| 184 | 14-3-3 | −1.0 | 1.1 | 1.5** | Bn | 224981577 | −13.7 | 22 | 4.8 | 35.2 | 4.77 | 28.91 |
| 189 | Guanine nucleotide-binding protein | 1.4* | −1.1 | −1.0 | Bn | 3023857 | −12.4 | 14 | 6.9 | 43.0 | 8.06 | 35.72 |
| 226 | Importin α | 1.2* | 1.0 | 1.3* | At | 238480717 | −8.3 | 9 | 5.1 | 67.9 | 5 | 58.91 |
Significant analysis of variance was followed by a Mann–Whitney test (p < 0.05) carried out on the normalized spot volumes (n = 6 for control; n = 4 for LS70, LS53, and LS32). Values indicate fold changes in protein abundance in LS seeds relative to control seeds in a linear scale. Significant differences from the control were at *p < 0.05 or **p < 0.01. The assigned best-matched protein is listed with the organism in which it was identified and its GenBank protein accession number. The log (E value), the percentage of sequence coverage (CV), and experimental (Exp.) and theoretical (Theo.) pI/Mr values obtained are also indicated.
Table VI. Significantly changed proteins in LS70, LS53, and LS32 mature seeds identified by mass spectrometry as involved in lipid storage and metabolism and in carbon metabolism and energy.
| Spot number | Protein name | LS70 | LS53 | LS32 | Species | NCBI accession number | log (E value) | CV (%) | Exp. pI | Exp. Mr (kDa) | Theo. pI | Theo. Mr (kDa) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lipid storage and metabolism | ||||||||||||
| 8 | Stereoleosin SLO1–2 | −1.4* | 1.3 | 3.3** | Bn | 196122096 | −19.7 | 26 | 6.7 | 51.7 | 6.26 | 39.11 |
| 19 | Caleosin | 1.3 | −1.3** | −2.3** | Bn | 196122112 | −5.3 | 11 | 5.9 | 31.0 | 5.81 | 28.13 |
| 59 | Stereoleosin SLO1–2 | 1.6** | −1.3 | −1.4 | Bn | 196122096 | −29.6 | 43 | 6.7 | 50.2 | 6.26 | 39.11 |
| 75 | Plastid-lipid associated protein | −1.1 | −1.4 | −2.1* | Br | 14248552 | −6.9 | 6 | 4.3 | 50.9 | 4.55 | 39.24 |
| 82 | Alcohol dehydrogenase | −1.2 | −1.3** | −2.1** | Br | 330414748 | −8.8 | 9 | 5.9 | 52.4 | 5.93 | 41.17 |
| 97 | Caleosin | 1.1 | −1.1 | −1.8** | Bn | 196122112 | −10.8 | 19 | 5.7 | 30.9 | 5.81 | 28.13 |
| 143 | Stereoleosin | 1.4* | 1.0 | −1.2 | Bn | 196122094 | −52.4 | 47 | 6.4 | 50.9 | 6.27 | 39.09 |
| 177 | Alcohol dehydrogenase | −1.5** | −1.4* | −1.4 | Br | 330414748 | −33.6 | 27 | 6.1 | 51.3 | 5.93 | 41.17 |
| 207 | Stereoleosin | −1.0 | −1.4* | −1.1 | Bn | 196122094 | −14.6 | 20 | 6.1 | 51.6 | 6.27 | 39.09 |
| 231 | β-ketoacyl-ACP synthetase 1 | −1.1 | −1.1* | −1.2 | Bn | 7385217 | −14.3 | 21 | 6.0 | 59.1 | 9.47 | 32.07 |
| 232 | Alcohol dehydrogenase Class 3 | 1.0 | −1.1 | −1.2* | At | 1143388 | −19.8 | 21 | 6.5 | 54.5 | 6.51 | 40.68 |
| Carbon metabolism and energy | ||||||||||||
| 16 | Succinate dehydrogenase | −2.1 | −3.1** | −1.8 | Al | 297797713 | −16.2 | 11 | 5.5 | 77.4 | 5.92 | 69.64 |
| 23 | Malate synthase, glyoxysomal | −1.2 | −1.8** | −3.0** | Bn | 126766 | −55.9 | 30 | 7.4 | 70.8 | 7.64 | 63.73 |
| 34 | Malate synthase, glyoxysomal | 1.0 | −1.8** | −2.7** | Bn | 126766 | −23.9 | 19 | 7.2 | 71.1 | 7.64 | 63.73 |
| 43 | UTP-glucose-1-phosphate uridylyltransferase | −2.5* | −1.7 | −2.2** | At | 15237947 | −10.9 | 14 | 5.5 | 64.0 | 5.72 | 51.92 |
| 44 | Phosphoglycerate kinase | −2.5** | −2.2* | −1.4 | At | 15219412 | −12.4 | 12 | 5.4 | 49.5 | 5.49 | 42.13 |
| 57 | Aconitase | 1.4* | 1.4* | 2.3** | At | 15224221 | −8.2 | 17 | 5.2 | 29.1 | 6.33 | 26.79 |
| 64 | Malate synthase, glyoxysomal | −1.0 | −1.7* | −2.3** | Bn | 126766 | −42.0 | 22 | 7.5 | 71.5 | 7.64 | 63.73 |
| 68 | Glucose and ribitol dehydrogenase | −1.5 | −1.0 | 1.5* | At | 75309952 | −37.4 | 35 | 5.9 | 36.4 | 6.11 | 31.39 |
| 74 | Transketolase | −1.3 | −1.5 | −2.2** | At | 18411711 | −17.9 | 10 | 5.4 | 82.9 | 5.94 | 79.97 |
| 81 | Glucose and ribitol dehydrogenase | −1.1 | 1.5** | 1.9** | At | 75309952 | −9.9 | 13 | 6.3 | 36.6 | 6.11 | 31.39 |
| 96 | Isocitrate dehydrogenase | −1.2 | 1.1 | 1.6** | At | 15218869 | −29.9 | 27 | 6.4 | 56.5 | 6.13 | 45.75 |
| 101 | Fructose-bisphosphate aldolase | −1.2 | −1.1 | 1.6** | At | 15231715 | −18.5 | 20 | 6.6 | 51.2 | 6.05 | 38.54 |
| 111 | Malate dehydrogenase, cytosolic | 1.2 | −1.2 | 1.5* | At | 21593565 | −9.1 | 11 | 6.5 | 49.3 | 7 | 35.66 |
| 120 | Phosphoglucose isomerase, cytosolic | 1.0 | 1.2 | 1.8** | At | 15239045 | −26.9 | 23 | 6.5 | 70.9 | 6.19 | 61.72 |
| 136 | Fructose-bisphosphate aldolase, cytosolic | 1.2 | −1.4** | −1.3 | Vv | 225440976 | −22.6 | 18 | 6.9 | 48.8 | 8.03 | 38.63 |
| 139 | NADP-malic enzyme | 1.3 | 1.7** | 1.5** | At | 15225262 | −6.3 | 5 | 6.8 | 74.0 | 6.32 | 64.28 |
| 148 | Aconitase | 1.2 | 1.0 | −1.4* | At | 15233349 | −45.6 | 23 | 6.0 | 92.5 | 5.98 | 98.15 |
| 165 | Glucose and ribitol dehydrogenase | −1.4** | 1.1 | 1.1 | At | 75309952 | −17.0 | 22 | 5.7 | 37.0 | 6.11 | 31.39 |
| 166 | Succinate-semialdehyde dehydrogenase | −1.0 | −1.5* | −1.3 | At | 15219379 | −27.7 | 24 | 6.1 | 63.6 | 6.51 | 56.56 |
| 171 | Glyceraldehyde-3-phosphate dehydrogenase | 1.0 | 1.1 | 1.5** | Bn | 310896467 | −26.4 | 38 | 6.3 | 49.8 | 6.81 | 15.69 |
| 181 | ATP synthase CF1 β subunit | −1.1 | −1.3* | −1.5** | At | 17939849 | −71.0 | 39 | 5.5 | 65.2 | 6.53 | 63.37 |
| 198 | Enolase | −1.2 | −1.2 | −1.4** | Br | 90194338 | −31.7 | 36 | 5.5 | 63.6 | 5.55 | 47.56 |
| 202 | ATP synthase CF1 β subunit | −1.3* | −1.3* | 1.1 | At | 17939849 | −96.5 | 54 | 5.4 | 66.2 | 6.53 | 63.37 |
| 205 | NADP-malic enzyme | 1.3** | 1.1 | 1.4** | At | 15225262 | −9.7 | 6 | 6.9 | 73.4 | 6.32 | 64.28 |
| 209 | V-type proton ATPase catalytic subunit A | 1.2* | 1.1 | −1.1 | Gm | 356521645 | −54.2 | 22 | 5.3 | 77.6 | 5.35 | 68.78 |
| 212 | Citrate synthase | 1.1 | 1.3* | 1.3* | At | 18406515 | −12.1 | 13 | 6.6 | 59.1 | 6.41 | 52.65 |
| 213 | Glyceraldehyde-3-phosphate dehydrogenase 2 | 1.1 | −1.3* | −1.1 | Bn | 241740186 | −44.6 | 40 | 6.7 | 48.6 | 7.7 | 36.94 |
| 216 | Phosphoglycerate kinase | 1.3* | 1.1 | 1.3 | At | 1022805 | −8.0 | 7 | 5.2 | 54.5 | 4.93 | 41.91 |
| 219 | NADP-malic enzyme | 1.2* | 1.2 | 1.3 | At | 15225262 | −18.6 | 18 | 6.9 | 73.5 | 6.32 | 64.28 |
| 228 | Enolase | −1.2** | 1.0 | 1.1 | Br | 34597330 | −85.3 | 56 | 5.6 | 64.1 | 5.46 | 47.38 |
| 230 | Malate dehydrogenase, cytosolic | −1.2 | −1.3* | −1.1 | At | 15239843 | −29.0 | 36 | 6.1 | 48.1 | 6.33 | 35.68 |
Significant analysis of variance was followed by a Mann–Whitney test (p < 0.05) carried out on the normalized spot volumes (n = 6 for control; n = 4 for LS70, LS53, and LS32). Values indicate fold changes in protein abundance in LS seeds relative to control seeds on a linear scale. Significant differences from the control were at *p < 0.05 or **p < 0.01. The assigned best-matched protein is listed with the organism in which it was identified and its GenBank protein accession number. The log (E value), the percentage of sequence coverage (CV), and experimental (Exp.) and theoretical (Theo.) pI/Mr values obtained are also indicated.
Table VII. Significantly changed proteins in LS70, LS53, and LS32 mature seeds identified by mass spectrometry as involved in stress response and as lectins.
| Spot number | Protein name | LS70 | LS53 | LS32 | Species | NCBI accession number | log (E value) | CV (%) | Exp. pI | Exp. Mr (kDa) | Theo. pI | Theo. Mr (kDa) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stress response | ||||||||||||
| 29 | Dehydroascorbate reductase | −1.8** | −1.1 | 1.6 | Br | 33285914 | −7.8 | 32 | 5.5 | 29.8 | 6.15 | 12.04 |
| 53 | Catalase | −2.4** | −1.9 | −1.7 | At | 1518450 | −12.4 | 9 | 7.1 | 61.6 | 6.95 | 56.99 |
| 56 | Mn superoxide dismutase | −2.1** | 1.1 | −1.2 | Eh | 148515008 | −19.8 | 21 | 6.6 | 28.0 | 8.77 | 25.50 |
| 66 | Peroxidase | −1.8** | 1.0 | 1.2* | Br | 253762012 | −16.4 | 24 | 5.0 | 55.2 | 6.34 | 35.37 |
| 113 | Cu/Zn superoxide dismutase | −1.1 | 1.2* | 1.7** | Rs | 2305111 | −8.3 | 16 | 5.5 | 14.6 | 5.44 | 15.10 |
| 122 | Glutathione S-transferase | −1.5** | −1.1 | 1.2* | Bo | 171921127 | −21.4 | 15 | 6.3 | 29.1 | 8.48 | 59.12 |
| 124 | Dehydroascorbate reductase | −1.4** | 1.1* | 1.3** | Br | 33285914 | −7.9 | 32 | 5.6 | 29.7 | 6.15 | 12.04 |
| 141 | Heat shock protein | −1.0 | −1.2 | −1.7** | Al | 297842179 | −45.7 | 23 | 6.2 | 92.9 | 5.86 | 101.20 |
| 144 | Fe superoxide dismutase | 1.3 | 1.4* | 1.7** | Bo | 334701491 | −10.9 | 24 | 6.1 | 26.9 | 5.97 | 23.90 |
| 145 | Cu/Zn superoxide dismutase | −1.1 | 1.3* | 1.5** | Rs | 2305111 | −10.4 | 16 | 5.9 | 14.8 | 5.44 | 15.10 |
| 162 | Heat shock protein 22 | 1.3* | −1.0 | −1.3 | Bn | 341872725 | −10.1 | 18 | 5.3 | 25.7 | 5.58 | 22.18 |
| 163 | Glyoxalase I | 1.3* | 1.6** | 1.5** | Bo | 2494843 | −8.9 | 22 | 5.5 | 36.1 | 5.16 | 31.65 |
| 164 | Glyoxalase I | 1.2** | 1.4** | 1.6** | Br | 157890952 | −4.3 | 14 | 5.2 | 35.9 | 5.35 | 31.91 |
| 169 | Glutathione S-transferase | −1.1 | −1.3* | −1.5** | Bj | 2204102 | −15.6 | 29 | 6.1 | 29.5 | 5.37 | 15.44 |
| 179 | Glyoxalase I | −1.4** | −1.2 | 1.1 | Bj | 3334244 | −7.9 | 25 | 5.3 | 27.1 | 5.56 | 20.78 |
| 185 | Catalase | −1.1 | −1.5** | −1.2 | Bn | 169244543 | −22.2 | 19 | 6.9 | 64.4 | 6.75 | 56.86 |
| 187 | Glyoxalase I | 1.0 | 1.3** | 1.5** | At | 15221116 | −7.0 | 9 | 5.4 | 35.3 | 5.17 | 31.93 |
| 201 | Glyoxalase I | 1.1 | 1.1 | 1.4* | Bo | 2494843 | −19.3 | 22 | 5.2 | 35.9 | 5.16 | 31.65 |
| 218 | Glutathione S-transferase | −1.3** | −1.3* | 1.0 | Bj | 170177802 | −65.2 | 42 | 6.5 | 28.9 | 5.81 | 22.64 |
| 220 | Peroxiredoxin antioxidant | −1.1 | −1.1 | −1.3** | Bn | 7381260 | −24.5 | 41 | 6.3 | 31.1 | 5.97 | 23.91 |
| 224 | Mn superoxide dismutase | −1.2** | 1.0 | 1.1 | At | 15228896 | −7.8 | 16 | 6.8 | 27.0 | 6.25 | 26.89 |
| 229 | Peroxiredoxin antioxidant | 1.1 | 1.1 | −1.2* | Bn | 7381260 | −45.0 | 45 | 5.7 | 30.7 | 5.97 | 23.91 |
| Lectins | ||||||||||||
| 80 | Jacaline like Jasmonate inductible protein | −1.3* | 1.2 | 1.6** | At | 34222076 | −20.7 | 8 | 5.9 | 36.5 | 6.03 | 49.28 |
| 196 | Jacaline like Jasmonate inductible protein | −1.5** | −1.3 | −1.2* | At | 34222076 | −16.5 | 10 | 6.2 | 62.2 | 6.03 | 49.28 |
Significant analysis of variance was followed by a Mann–Whitney test (p < 0.05) carried out on the normalized spot volumes (n = 6 for control; n = 4 for LS70, LS53, and LS32). Values indicate fold changes in protein abundance in LS seeds relative to control seeds on a linear scale. Significant differences from the control were at *p < 0.05 or **p < 0.01. The assigned best-matched protein is listed with the organism in which it was identified and its GenBank protein accession number. The log (E value), the percentage of sequence coverage (CV), and experimental (Exp.) and theoretical (Theo.) pI/Mr values obtained are also indicated.
Modulations of SSPs, Proteins Involved in Protein Synthesis and Processing, and Late Embryogenesis Abundant Proteins
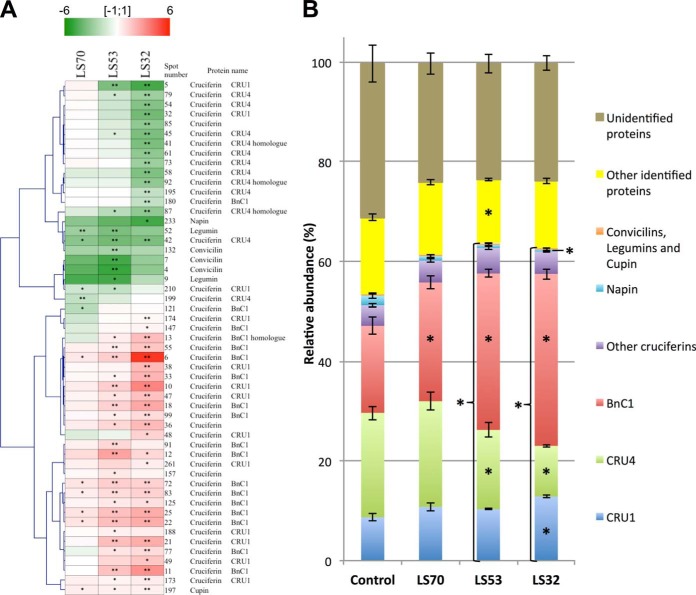
The proteomic analysis of mature seeds revealed that S limitation led to significant modulations of SSP accumulation (Table IV). The number of changed SSP spots increased with the precocity of S deficiency: only 11 SSP spots were affected by LS70 treatment, versus 42 SSP spots for LS32 treatment (Fig. 4B, Fig. 5A, Table IV). LS70 seeds showed a significant accumulation of SSP spots identified as BnC1 subunits (about +1.5-fold), which are relatively sparse in cysteine and methionine (representing 1.84% of total amino acids in this protein). In contrast, the relative abundance of protein spots identified as cruciferin Cru4 subunits, an S-rich SSP (S amino acids represent 2.80% of the total amino acids), decreased in LS70 seeds. LS53 seeds were also characterized by an accumulation of BnC1 and a decrease in Cru4 abundance, but these changes were more pronounced than in LS70 seeds. Markedly, early S limitation (LS32 treatment) drastically affected Cru4 accumulation and significantly increased the abundance of SSP spots that corresponded to BnC1. Surprisingly, the majority of protein spots corresponding to an S-rich SSP (Cru1, 3.34% S amino acids in total amino acids) increased in abundance in LS53 and LS32 seeds relative to the control. LS53 and LS32 treatments also led to an accumulation of low-Mr spots corresponding to BnC1 (spots 11 and 77) and Cru1 (spots 21 and 49) in mature seeds.
Fig. 5.
Adaptation of seed storage protein (SSP) accumulation in response to LS70, LS53, and LS32 treatments. A, clustering of expression profiles of significantly modulated SSPs identified by mass spectrometry in LS70, LS53, and LS32 mature seeds relative to the control (n = 6 for control; n = 4 for LS70, LS53, and LS32). Proteins up- and down-regulated in the whole seed are marked in red and green, respectively (color code at the top of the figure). B, relative abundance of the different classes of SSPs and other proteins in the total protein content of Brassica napus mature seeds grown under control, LS70, LS53, and LS32 conditions. The values correspond to the mean ± S.E. (n = 6 for control; n = 4 for LS70, LS53, and LS32). Significant differences from the control were at *p < 0.05 or at **p < 0.01.
In order to compare the contribution of the different SSPs to the total protein pool depending on the treatment, the proportion of each SSP in the total protein content was calculated (Fig. 5B). Interestingly, the global contribution of SSPs to the total protein content was significantly increased in LS53 and LS32 seeds relative to the control. This increase was principally associated with a greater relative accumulation of BnC1 in LS53 seeds and BnC1 and Cru1 in LS32 seeds, whereas the accumulation of Cru4 was reduced in LS53 and LS32 seeds relative to the control (Fig. 5B). Considering the percentage of S amino acids in each SSP and in the other proteins, the theoretical proportions of S amino acids found in total proteins from control and S-limited seeds were also calculated (Fig. 6A). This proportion was significantly decreased in LS70, LS53, and LS32 seeds relative to the control. In accordance with these calculations, the S proteic content measured by mass spectrometry after soluble protein extraction and purification was significantly less in LS70, LS53, and LS32 seeds than in control seeds (up to 49% for LS32 treatment; Fig. 6B). However, the S proteic content, which accounted for 51.3% of total S in control seeds, respectively represented 59.9%, 57.9%, and 80.9% of the total S in LS70, LS53, and LS32 seeds (Fig. 6B).
Fig. 6.
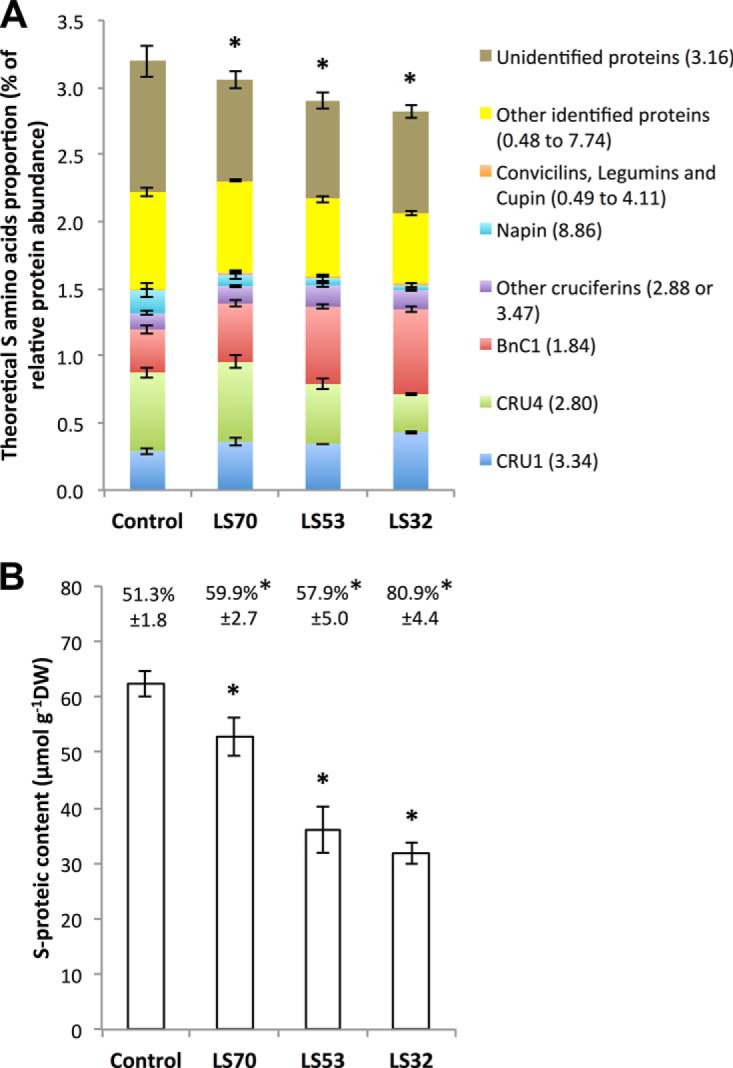
Effect of control, LS70, LS53, and LS32 treatments on theoretical and experimentally measured sulfur contents of mature seed proteins. A, theoretical S amino acid proportion calculated as the sum of relative abundances multiplied by the S amino acids (cysteine and methionine) proportion of the corresponding protein, given next to the protein name in brackets. B, S-proteic content of mature seeds grown under control, LS70, LS53, or LS32 conditions. For each treatment, the S-proteic/S ratio is shown directly above the corresponding bar. In both panels the values correspond to the mean ± S.E. (n = 6 for control; n = 4 for LS70, LS53, and LS32). *Significant difference from the control value (p < 0.05).
Among proteins with a function in protein synthesis, proteomic analysis revealed a reduction of translation initiation factor accumulation (spots 108, 110, and 142) in LS70 and LS53 seeds relative to the control, and a spot corresponding to elongation factor 1α (spot 100) was less abundant in LS32 seeds. In contrast, spot 110 was increased in LS32 seeds relative to the control (1.3-fold). An accumulation of two protein spots identified as 60S acidic ribosomal protein (spots 194 and 217) was observed in LS32 seeds, whereas the relative abundance of a chaperonin (spot 140) and a glycosyl hydrolase family 38 protein (spot 168), both involved in protein processing, decreased in LS32 seeds (Table IV).
Sulfur limitation also led to the differential accumulation of numerous protein spots identified as late embryogenesis abundant proteins. Among them, three spots very close to a late embryogenesis abundant protein from Arabidopsis thaliana (spots 182, 191, and 225; gi:15231736) specifically expressed during seed ripening (The Arabidopsis Information Resource) increased significantly in LS seeds (Table IV).
Proteome Changes Related to S and N Metabolism, Cell Structure, Growth, and Signal Transduction
In our experiment, the protein spots corresponding to myrosinase were significantly reduced in LS53 and LS32 mature seeds, whereas a differential accumulation of myrosinase-binding proteins (MBPs) (Table V) according to the period of S deficiency was revealed. The thiamine biosynthetic enzyme THI1 decreased significantly in LS53 and LS32 seeds relative to the control (spot 40, Table V). This enzyme is involved in thiamine synthesis from glyceraldehyde-3-phosphate and cysteine. Thiamine is the precursor of the coenzyme thiamine pyrophosphate, required for the activity of some decarboxylases such as pyruvate decarboxylase, pyruvate dehydrogenase, pyruvate oxidase, and transketolase (31). THI1 is also suggested to be involved in mitochondrial DNA damage tolerance in plant cells (32).
Although secondary S metabolism appears to be highly modulated in seeds subjected to S deficiency, only weak changes associated with primary S metabolism were observed: two protein spots corresponding to methionine synthase were significantly less abundant in LS53 and LS32 seeds than in the control (spots 158 and 176, Table V).
A protein spot related to N metabolism identified as a glycine cleavage T-protein, involved in glycine conversion to serine in the course of photorespiration, was significantly reduced in LS32 seeds relative to the control (spot 37, Table V). We also observed that the three S limitation treatments led to an increased accumulation of β-tubulin in mature seeds (spots 160, 161, and 214; Table V) and to differential accumulation of actin isoforms. The abundance of 51.7-kDa actin (spot 95) was significantly reduced in LS32 mature seeds. This protein is homologous to actin 7 from A. thaliana (99% similarity, At5G09810, gi:15242516; The Arabidopsis Information Resource) whose mutants are defective in germination and root growth (BLASTp from NCBI). Conversely, LS32 seeds showed an accumulation of a 24.8-kDa actin (spot 152) homologous to actin 11 from A. thaliana (91% similarity, At3G12110, gi:15229955; The Arabidopsis Information Resource) expressed predominantly during reproductive development.
The proteomic analysis also revealed changes in the expression of some proteins involved in signal transduction: a guanine nucleotide-binding protein (spot 189) and an importin α (spot 226) both accumulated in LS70 seeds relative to the control. A 14–3-3 protein (spot 184) was also accumulated in LS32 seeds relative to control seeds (Table V).
Modulation of Proteins Involved in Lipid Storage, C metabolism, and Energy
Different classes of proteins associated with lipid storage were modulated in LS seeds (Table VI). The abundance of two protein spots corresponding to caleosins (spots 19 and 97), a minor group of integral lipid body proteins able to bind Ca2+ (33), was decreased in LS53 and, even more so, in LS32 seeds, but these protein spots were not significantly affected by LS70 treatment (Table VI). Nevertheless, protein spots corresponding to stereoleosins did not exhibit the same patterns and were differentially regulated by S restriction treatments.
The three S limitation treatments applied in our study led to substantial modulations of numerous spots related to C metabolism, as depicted in Fig. 7 (also see Table VI). LS70 seeds showed a reduction of UDP-glucose pyrophosphorylase (spot 43), phosphoglycerate kinase (spot 44), enolase (spot 228), alcohol dehydrogenase (spot 177), and the β-subunit of mitochondrial F1 ATP synthase (spot 202). In contrast, the abundance of aconitase (spot 57) and malic enzymes (spots 205 and 219) increased in LS70 and LS53 seeds relative to the control. In LS53 seeds, there was also an accumulation of β-glucosidases (spots 14, 170, and 211) and a reduction of glyceraldehyde-3-phosphate dehydrogenase 2 (GAPDH) (spot 213), succinate dehydrogenase (spot 16), cytosolic MDH (spot 230), and glyoxysomal malate synthase (spots 23, 34, and 64). Interestingly, as for LS53 seeds, LS32 seeds showed an overaccumulation of citrate synthase (spot 212), aconitase (spots 57 and 148), and isocitrate dehydrogenase (spot 96), the first TCA cycle enzymes, and reduced abundance of glyoxysomal malate synthase (spots 23, 34, and 64). In LS32 seeds, there was also an accumulation of phosphoglucose isomerase (spot 120), fructose bisphosphate aldolase (spot 101), and GAPDH (spot 171), three enzymes involved in the first steps of glycolysis. Cytosolic MDH (spot 111) was also overaccumulated in LS32 seeds (about +1.5-fold) relative to the control.
Fig. 7.
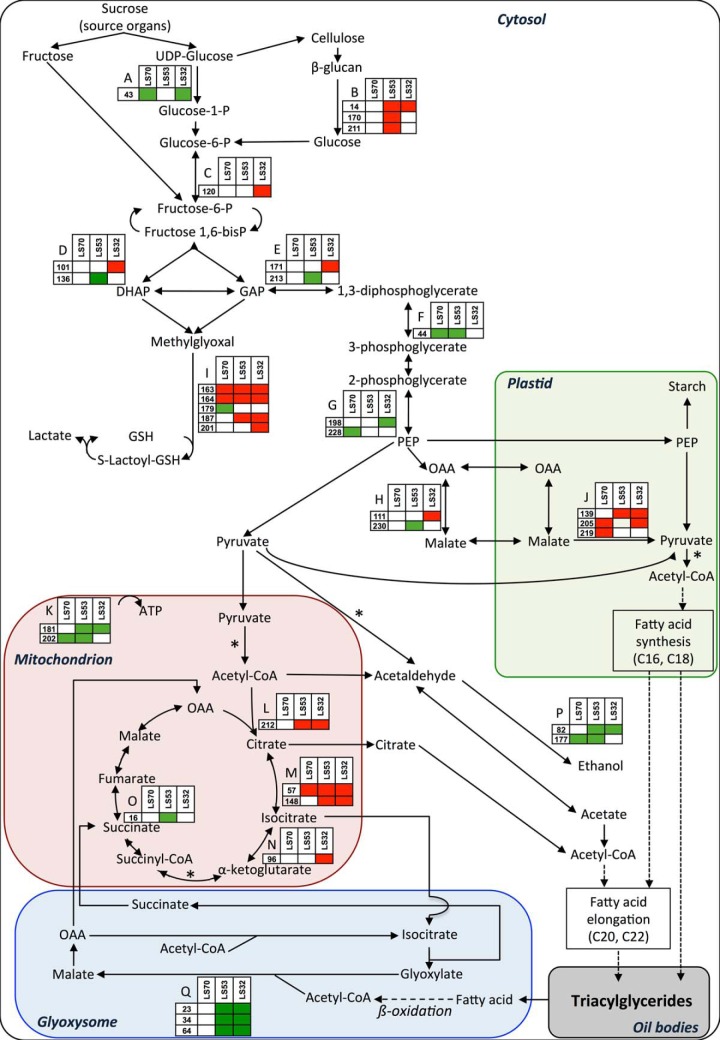
Schematic representation of changes in accumulation of proteins of carbon metabolism and associated proteins in mature LS70, LS53, and LS32 seeds compared with control seeds. Significantly up- and down-regulated proteins are shown in red and green, respectively. *Thiamine-pyrophosphate-dependent reaction. A, UTP-glucose-1-phosphate uridylyltransferase. B, β-glucosidase. C, cytosolic phosphoglucose isomerase. D, fructose-bisphosphate aldolase. E, glyceraldehyde-3-phosphate dehydrogenase. F, phosphoglycerate kinase. G, enolase. H, cytosolic malate dehydrogenase. I, glyoxalase I. J, NADP-malic enzyme. K, ATP synthase CF1 β subunit. L, citrate synthase. M, aconitase. N, isocitrate dehydrogenase. O, succinate dehydrogenase. P, alcohol dehydrogenase. Q, glyoxysomal malate synthase. DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde 3-phosphate; GSH, reduced form of glutathione; OAA, oxaloacetate.
Interestingly, our proteomic analysis revealed significant accumulation of protein spots identified as glyoxalase I (lactoylglutathione lyase; spots 163, 164, 187, and 201; Fig. 7) with the three treatments applied, except for one spot over three in LS70 seeds that was increased (spot 179). This protein, along with a reduced form of glutathione, is involved in methylglyoxal detoxification formed from nonenzymatic phosphate elimination of glyceraldehyde-3-phosphate or dihydroxyacetone phosphate (34).
Modulation of Proteins Involved in Plant Stress Response and Lectins
In LS70 and LS53 seeds, there was decreased accumulation of catalase (spots 53 and 185) and glutathione-S-transferase (spots 122, 169, and 218; Table VII). Interestingly, a decrease in Mn-superoxide dismutase (SOD) (spots 56 and 224) and dehydroascorbate reductase (spots 29 and 124) was also observed in LS70 seeds relative to the control. Conversely, LS53 and LS32 seeds showed an overaccumulation of Cu/Zn-SOD (spots 113 and 145). As shown in Table VII, two spots corresponding to jacalin-like jasmonate inducible proteins were significantly repressed in LS70 seeds relative to the control.
DISCUSSION
This study clearly demonstrated that the effect of S limitation on components of seed yield depend on the timing of the deficiency during the ontogenetic cycle of B. napus. Previous research emphasized the crucial role of leaves as the major source organ for S in response to S restriction (4). Despite enhancement of the S remobilization processes, the LS70, LS53, and LS32 treatments led to a reduction of the S content in mature seeds, which could explain the reduced accumulation of S in proteins, GLS, and glutathione (Tables I and III).
Late S deficiency (applied at the start of pod filling (i.e. LS70 treatment)) did not affect seed yield or oil, lipid, and protein contents, but it did lead to reduced seed protein quality and GLS content. Severe decreases in seed yield and/or protein and lipid quality were observed when S limitation occurred at early stages of development (bolting stage (LS32) or visible bud/early flowering stage (LS53)). A negative effect on seed yield was also observed in Medicago truncatula (22) in response to S deficiency from the mid-vegetative stage and was related to a shift in carbon and nitrogen allocation patterns in favor of the root system. In oilseed rape, the reductions in seed yield and nutritional quality in response to S restriction were associated with specific physiological and proteomics changes (Fig. 7).
Effects of Early S Limitation on Seed Oil Content and Quality Are Related to Disturbances of C Metabolism and Energy
The nutritional value and usefulness of vegetable oils depend on their fatty acid balance. Whereas a late S limitation (LS70) did not result in a reduction in oil content and quality, LS53 and LS32 treatments led to reduced accumulation of fatty acids in seeds, mainly due to a decrease in C18 derivatives, especially the two essential fatty acids, linoleic (ω6) and linolenic (ω3) acid (Table IV). These results are consistent with the results of a study conducted on Brassica campestris (35). The significant decrease of the lipid-body-associated proteins caleosins in LS53 and LS32 seeds (Table VI) is in accordance with the lower lipid amount of these seeds. In contrast, the relative maintenance of these caleosins and the induction of stereoleosins in LS70 seeds could participate in the conservation of a high lipid amount in these seeds.
Interestingly, the lower fatty acid content in LS53 and LS32 seeds is mainly associated with a lesser amount of C18 but not of C20 and C22 fatty acids, suggesting that S limitation leads to a decrease in fatty acid synthesis, although their elongation to very long chains does not seem to be affected (Fig. 2). Although the present proteomic analysis was performed on mature seeds, this hypothesis is consistent with the characterization of changes in proteins associated with C metabolism, strongly suggesting that glycolysis and the TCA cycle are affected in developing seeds under S limitation. For example, the significant decreases in phosphoglycerate kinase and enolase (Fig. 7) might be a source of disturbance in C flux through glycolysis that might contribute to the reduced fatty acid content observed in LS53 and LS32 seeds. Also, the results suggest a redirection of the glycolic flux toward methylglyoxal metabolism previously shown to be activated by various abiotic stresses (36). It is worth noting in this context that the abundance of chloroplastidial malic enzyme is up-regulated by S deficiency, suggesting that the PEP carboxylase–MDH–malic enzyme pathway is induced under S-limited conditions and that its contribution to the acetyl-CoA supply for fatty acid synthesis is substantial. Moreover, the data revealed a reduction of malate synthase, a specific enzyme of the glyoxylate cycle involved in acetyl-CoA conversion to carbohydrates (37), which might have resulted in reduced C fluxes through the glyoxylate cycle in LS53 and LS32 seeds. This could allow a down-regulation of fatty acid β-oxidation that might help to keep the oil content as high as possible, although these metabolic adjustments are not sufficient for LS53 and LS32 treatments. In addition, the accumulation of citrate synthase, aconitase, and isocitrate dehydrogenase suggests that citrate formation in mitochondria is promoted, which might allow the maintenance of cytosolic fatty acid elongation to very long chains. Interestingly, among the proteins newly identified as variables in response to S limitation is the thiazole biosynthetic enzyme THI1 (spot 40). The low abundance of THI1 observed in LS53 and LS32 seeds might result in a decline in the synthesis of thiamine that could impact negatively on carbon metabolism and thus on fatty acid synthesis. Indeed, thiamine is the precursor of thiamine pyrophosphate, an essential coenzyme for the activity of several enzymes associated with primary metabolism such as pyruvate decarboxylase and pyruvate dehydrogenase, two enzymes strongly involved in the synthesis of acetyl-CoA that could be used in the TCA cycle and/or fatty acid elongation (Fig. 7).
The repression of succinate dehydrogenase, directly involved in the transport chain of the inner mitochondrial membrane electron, and the repression of the ATP synthase β-subunit strongly suggest that energy production was diminished in S-limited seeds relative to the control. Although mitochondrial ATP production contributes to only a small part of the energy required for seed development (9), this repression could also explain the reduced accumulation of storage compounds. Together, these proteomics changes provide insights into processes that might contribute to the reduction of fatty acid accumulation and to the alteration of lipid composition (in favor of fatty acids with long chains) observed in LS53 and LS32 seeds.
Seed Metabolic Changes Induced in Response to S Limitation Might Contribute to the Maintenance of Protein Content but Not Quality
Meal quality would be reduced if oil was extracted from seeds obtained from early S-limited plants. Indeed, data obtained via theoretical calculation from proteomic analysis (Fig. 6A) and mass spectrometry (Fig. 6B) revealed that the reduced accumulation of S-rich SSPs in response to S limitation resulted in a lower S-proteic content than in controls. However, the seed protein content remained high after the LS32 and LS53 treatments (Table I). Thus, under S limitation, the protein content of B. napus seeds can be maintained at a level very close to that of controls by decreasing the S-rich/S-poor protein ratio, as previously demonstrated in Arabidopsis, lupin, and pea (38–40). However, this adaptation strategy is not sufficient for LS32 treatment, in which the seed protein content is affected, as observed in Arabidopsis thaliana (14), maybe as a result of a reduction of N remobilization from leaves, as previously reported by Dubousset et al. (4), and/or N assimilation in seeds.
The decreased level of Cru4 (S-rich cruciferin) and the increased level of BnC1 (S-poor cruciferin) strongly indicate that SSP balance depends on S amino acid availability in developing seeds. In contrast, the increased abundance of Cru1 (S-rich cruciferin) in S-limited seeds might help to maintain a reasonable amount of S in the form of proteins and suggests a specific fine-tuning of oilseed rape SSP expression (Table IV, Figs. 5A and 5B). Interestingly, whereas the reduced accumulation of translational initiation factor (spots 108, 110, and 142) in LS70 and LS53 seeds suggests a decrease in protein synthesis, LS32 treatment caused a repression of spots related to protein elongation and degradation.
Seed transcriptome analysis revealed that genes involved in S metabolism are expressed in B. napus seeds (41), and our recent work shows that S assimilation is functional in this organ (42). The present study might suggest that sulfate assimilation during seed development is enhanced by S limitation, as the S-sulfate/S ratio was strongly reduced in LS53 and LS32 seeds and the S-proteic/S ratio was significantly greater in LS70, LS53, and LS32 seeds than in the control (Table III, Fig. 6B). However, these observations could also be the result of an important metabolic sink strength for sulfur related to protein synthesis. Although the free cysteine content remained very low, it was significantly higher in LS53 and LS32 seeds than in the control (Table III), which might suggest that cysteine availability is not limiting for S-rich SSP accumulation. However, this unexpected result might rather be related to a defect in cysteine incorporation into proteins during the final stages of seed development. In contrast, the repression of methionine synthase that could reduce methionine synthesis might result in a limitation of S-rich SSP accumulation.
Proteins Involved in Plant Stress Response Exhibit Different Responses Depending on the Severity of S Limitation
In contrast to Arabidopsis, B. napus seeds show high myrosinase activity, consisting in the cleavage of GLS to aglucons and glucose. Depending on different parameters (i.e. the pH or the side chain of GLS), the aglucons decompose to form toxic products such as isothiocyanates, thiocyanates, nitriles, or epithionitriles, involved in defense mechanisms against pathogens. Myrosinases are specifically expressed in the vacuoles of myrosin cells in B. napus embryo, whereas the different myrosinase-binding proteins that regulate myrosinase activity are present in other cell types (43). Surprisingly, the low myrosinase abundance observed in LS53 and LS32 seeds suggests that GLS degradation is potentially reduced in seeds subjected to S limitation. Even though the cultivar studied is a double low oilseed rape, GLS accumulation still strongly decreased in S-limited seeds, indicating that the GLS content in these seeds is principally regulated by their synthesis in pod walls during seed development, probably through a reduction in S supply and the regulation of S assimilation. The inhibition of myrosinase also highlights the importance of maintaining GLS in mature seeds, probably for its role in defense against pathogens, as demonstrated in leaves (44), which could be significant during germination or seedling establishment. We could then assume that S-limited seeds, particularly LS53 and LS32 seeds, are more susceptible to pathogen attacks. Similar to the low GLS content in S-limited seeds, the repression of the thiazole biosynthetic enzyme THI1 could reduce the resistance of these seeds to pathogen attacks, as thiamine was shown to induce systemic acquired resistance in plants (45) and indicates that the secondary S metabolism is reduced under S limitation. Although the adaptation mechanisms are still unknown, our data suggest the involvement of a demand-driven control of sulfate assimilation to sustain protein accumulation through S amino acid synthesis rather than secondary S compounds such as thiamine, acetyl-CoA, and GLS.
The modulations of stress-response-associated proteins revealed different responses of the seeds to the three S limitation treatments. LS70 seeds exhibited reduced expression of dehydroascorbate reductase and Mn-SOD, whereas LS53 and LS32 seeds showed accumulation of dehydroascorbate reductase and, particularly, Cu/Zn-SOD, as previously observed in B. napus young leaves (21) and A. thaliana seeds (14). These inductions might be a response to oxidative stress due to a diminished amount of glutathione under low S availability. Similarly, to compensate for the low glutathione content that conducts to a lower capacity to detoxify harmful molecules produced during seed development such as methylglyoxal, an induction of glyoxalase I was observed in S-limited seeds. The accumulation of glyoxylase I seen in LS53 and LS32 seeds could facilitate methylglyoxal detoxification despite the decrease in glutathione (Fig. 7). This could be particularly important in LS32 seeds, in which the reduced accumulation of fatty acids and the induction of β-glucosidase, phosphoglucose isomerase, and fructose bisphosphate aldolase suggest an accumulation of carbohydrates such as glyceraldehyde-3-phosphate or dihydroxyacetone phosphate that could increase nonenzymatic methylglyoxal formation, as previously shown in A. thaliana leaves (46). Interestingly, the maintenance of glutathione content in LS70 seeds and the reduced accumulation of dehydroascorbate reductase and Mn-SOD suggest that glutathione may be efficiently used for reactive oxygen species detoxification as in control seeds.
The Metabolic Changes Caused by Severe S Deficiency Lead to Defects in Germination
The present study highlights the finding that germination speed and the final germination rate were not significantly affected in seeds from Brassica napus subjected to S limitation applied at early flowering (LS53) or the start of pod filling (LS70). This contrasts with data obtained in Medicago truncatula (22) showing a reduced germination speed or a reduced final percentage of germinated seeds when the mother plants were S-limited at the start of seed filling or at flowering, respectively. One characteristic of LS seeds in common between Medicago truncatula and Brassica napus is the delay in the start of germination when the mother plants are S-limited at or before flowering. This could be attributed to a lower abundance in mature seeds of enzymes involved in carbon metabolism and energy (see Table VI and Ref. 22) that are presumably important for the rapid switch from quiescence to active metabolism during imbibition. A severe S restriction (LS32 treatment) results in the decline of Brassica napus seed vigor (Table II), which is not surprising considering the involvement of S metabolism in seed germination (47). Interestingly, for LS32, the percentage of viable seeds was quite similar to the percentage of germinated seeds, meaning that the nongerminated seeds were probably nonviable. Recent work in Medicago truncatula (22) has also shown that the seeds produced from mother plants subjected to S limitation from the mid-vegetative stage present a significantly slower germination speed. In Medicago truncatula, these results could be explained by a reduced level of sucrose and raffinose family oligosaccharides in mature seeds (22). In LS seeds of Brassica napus, the presence of late embryogenesis abundant proteins that are specific of late maturation (spots 182, 191, and 225), the low water content, and the brown color of the seeds might suggest that late maturation events occurred. However, some processes that are characteristic of proper maturation appeared to be affected in LS seeds, such as lipid content (Table I) and the switch from oil accumulation to oil degradation, evidenced by the greater accumulation of malic enzyme and proteins involved in the Krebs cycle and the reduced accumulation of glyoxysomal malate synthase. The low abundance of the latter enzyme, which is essential for the use of fatty acids during seedling establishment, could also result in a default in these critical stages of development (37).
Although these changes were more pronounced in response to LS32 treatment, they are not specific to LS32 seeds and cannot therefore be the sole explanation for their reduced viability. In LS32 mature seeds, the lower abundance of 51.7-kDa actin, homologous to actin 7 from A. thaliana (spot 95), and the accumulation of a 24.8-kDa actin homologous to actin 11 from A. thaliana (spot 152) highlight specific effects of this early S restriction treatment on seed maturation that could be associated with the seeds' reduced viability. Other proteomic changes, such as the modulations of β-tubulin and β-glucosidase, which could affect cellular development, or the reduction of myrosinase and GLS accumulation, which could prevent a good defense against pests, are also potentially negative for efficient germination, seedling establishment, and growth.
If S reserves are sufficient, B. napus seeds are able to employ specific metabolic compensations to maintain seed yield and/or quality in the case of S limitation. Moreover, the proteome study revealed metabolic processes that could contribute to the reduction of fatty acid accumulation and to the alteration of lipid composition in S-limited plants. Further investigations will be necessary (i) to study the effect of S limitation on seed development and (ii) to verify whether the viable seeds produced by LS plants are more susceptible to biotic/abiotic stress and/or an alteration of seedling development.
Supplementary Material
Acknowledgments
We thank Mrs. Nathalie Nesi and Véronique Gautier (IGEEP, INRA Rennes, France) for their help in seed composition analysis, M. Julien Besnard (UMR EVA, Caen, France) for the obtention of germination data, Mrs. Delphine Aimé (INRA Dijon, France) for her assistance in the achievement of two-dimensional gel electrophoresis, Mr. Laurent Coquet (Plateforme de protéomique, Rouen, France) for LC/MS-MS analyses, and Mr. Olivier Langella (PAPPSO, UMR de Génétique Végétale, INRA Gif-sur-Yvette, France) for PROTICdb assistance.
Footnotes
Author contributions: P.D., L.D., J.A., and J.T. designed research; P.D., L.D., K.G., S.K., J.A., and J.T. performed research; P.D. and K.G. contributed new reagents or analytic tools; P.D., L.D., K.G., S.K., J.A., and J.T. analyzed data; P.D., K.G., J.A., and J.T. wrote the paper.
* This work was supported by the national SERAPIS program (Optimization of the Sulphur Fertilization: Development of Innovative Fertilizers and Indicators of Sulphur Nutrition in Plants). The SERAPIS program is coordinated by the private company TIMAC AGRO INTERNATIONAL–ROULLIER Group and benefits from financial support from the Région Basse-Normandie, Région Bretagne, Région Pays-de-la-Loire, FEDER Basse-Normandie, Fonds Unique Interministériel, Roullier, Angers Loire Métropole. This work was also supported by a Ph.D. grant to Philippe D'Hooghe from the French Ministry of Research.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PEP
- phosphoenolpyruvate
- ATP
- adenosine triphosphate
- GLS
- glucosinolate
- LS
- low sulfur
- MDH
- malate dehydrogenase
- DW
- dry weight
- Mr
- molecular weight
- SOD
- superoxide dismutase
- SSP
- seed storage protein
- TCA
- trichloroacetic acid
- THI
- thiazole biosynthetic enzyme.
REFERENCES
- 1. McGrath S. P., Zhao F.-J. (1996) Sulphur uptake, yield responses and the interactions between nitrogen and sulphur in winter oilseed rape (Brassica napus). J. Agr. Sci. 126, 53–62 [Google Scholar]
- 2. Zhao F.-J., Bilsborrow P. E., Evans E. J., McGrath S. P. (1997) Nitrogen to sulphur ratio in rapeseed and in rapeseed protein and its use in diagnosing sulphur deficiency. J. Plant Nutr. 20, 549–558 [Google Scholar]
- 3. Scherer H. W. (2001) Sulphur in crop production—invited paper. Eur. J. Agron. 14, 81–111 [Google Scholar]
- 4. Dubousset L., Etienne P., Avice J.-C. (2010) Is the remobilization of S and N reserves for seed filling of winter oilseed rape modulated by sulphate restrictions occurring at different growth stages? J. Exp. Bot. 61, 4313–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eastmond P. J., Rawsthorne S. (2000) Coordinate changes in carbon partitioning and plastidial metabolism during the development of oilseed rape embryos. Plant Physiol. 122, 767–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schwender J., Ohlrogge J. B., Shachar-Hill Y. (2003) A flux model of glycolysis and the oxidative pentosephosphate pathway in developing Brassica napus embryos. J. Biol. Chem. 278, 29442–29453 [DOI] [PubMed] [Google Scholar]
- 7. Alban C., Job D., Douce R. (2000) Biotin metabolism in plants. Annu. Rev. Plant. Biol. 51, 17–47 [DOI] [PubMed] [Google Scholar]
- 8. Plaxton W. C. (1996) The organization and regulation of plant glycolysis. Annu. Rev. Plant. Biol. 47, 185–214 [DOI] [PubMed] [Google Scholar]
- 9. Schwender J., Shachar-Hill Y. (2006) Mitochondrial metabolism in developing embryos of Brassica napus. J. Biol. Chem. 281, 34040–34047 [DOI] [PubMed] [Google Scholar]
- 10. Baud S., Lepiniec L. (2010) Physiological and developmental regulation of seed oil production. Prog. Lipid Res. 49, 235–249 [DOI] [PubMed] [Google Scholar]
- 11. Bewley J. D., Black M. (1994) Seeds: Physiology of Development and Germination (2nd Ed.), Plenum Press, New York [Google Scholar]
- 12. Gallardo K., Job C., Groot S. P., Puype M., Demol H., Vandekerckhove J., Job D. (2001) Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol. 126, 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gallardo K., Job C., Groot S. P., Puype M., Demol H., Vandekerckhove J., Job D. (2002) Proteomics of Arabidopsis seed germination. A comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol. 129, 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Higashi Y., Hirai M. Y., Fujiwara T., Naito S., Noji M., Saito K. (2006) Proteomic and transcriptomic analysis of Arabidopsis seeds: molecular evidence for successive processing of seed proteins and its implication in the stress response to sulfur nutrition. Plant J. 48, 557–571 [DOI] [PubMed] [Google Scholar]
- 15. Ufaz S., Galili G. (2008) Improving the content of essential amino acids in crop plants: goals and opportunities. Plant Physiol. 147, 954–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Altenbach S. B., Kuo C. C., Staraci L. C., Pearson K. W., Wainwright C., Georgescu A., Townsend J. (1992) Accumulation of a Brazil nut albumin in seeds of transgenic canola results in enhanced levels of seed protein methionine. Plant Mol. Biol. 18, 235–245 [DOI] [PubMed] [Google Scholar]
- 17. Pastorello E. A., Pompei C., Pravettoni V., Brenna O., Farioli L., Trambaioli C., Conti A. (2001) Lipid transfer proteins and 2S albumins as allergens. Allergy 5, 45–47 [DOI] [PubMed] [Google Scholar]
- 18. Lee M., Huang T., Toro Ramos T., Fraga M., Last R. L., Jander G. (2008) Reduced activity of Arabidopsis thaliana HMT2, a methionine biosynthetic enzyme, increases seed methionine content. Plant J. 54, 310–320 [DOI] [PubMed] [Google Scholar]
- 19. Kopriva S., Rennenberg H. (2004) Control of sulphate assimilation and glutathione synthesis: interaction with N and C metabolism. J. Exp. Bot. 55, 1831–1842 [DOI] [PubMed] [Google Scholar]
- 20. Hirai M. Y., Yano M., Goodenowe D. B., Kanaya S., Kimura T., Awazuhara M., Arita M., Fujiwara T., Saito K. (2004) Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 101, 10205–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. D'Hooghe P., Escamez S., Trouverie J., Avice J.-C. (2013) Sulphur limitation provokes physiological and leaf proteome changes in oilseed rape that lead to perturbation of sulphur, carbon and oxidative metabolisms. BMC Plant Biol. 13, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zuber H., Poignavent G., Le Signor C., Aimé D., Vieren E., Tadla C., Lugan R., Belghazi M., Labas V., Santoni A.-L., Wipf D., Buitink J., Avice J.-C., Salon C., Gallardo K. (2013) Legume adaptation to sulfur deficiency revealed by comparing nutrient allocation and seed traits in Medicago truncatula. Plant J. 76, 982–996 [DOI] [PubMed] [Google Scholar]
- 23. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 24. Koprivova A., North K. A., Kopriva S. (2008) Complex signaling network in regulation of adenosine 5′-phosphosulfate reductase by salt stress in Arabidopsis roots. Plant Physiol. 146, 1408–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gallardo K., Firnhaber C., Zuber H., Héricher D. (2007) A combined proteome and transcriptome analysis of developing Medicago truncatula seeds. Mol. Cell. Proteomics 6, 2165–2179 [DOI] [PubMed] [Google Scholar]
- 26. Mathesius U., Keijzers G., Natera S. H., Weinman J. J., Djordjevic M. A., Rolfe B. G. (2001) Establishment of a root proteome reference map for the model legume Medicago truncatula using the expressed sequence tag database for peptide mass fingerprinting. Proteomics 1, 1424–1440 [DOI] [PubMed] [Google Scholar]
- 27. Ferry-Dumazet H., Houel G., Montalent P., Moreau L., Langella O., Negroni L., Vincent D., Lalanne C., de Daruvar A., Plomion C., Zivy M., Joets J. (2005) PROTICdb: a web-based application to store, track, query, and compare plant proteome data. Proteomics 5, 2069–2081 [DOI] [PubMed] [Google Scholar]
- 28. Brown R. F., Mayer D. G. (1988) Representing cumulative germination. 2. The use of the Weibull function and other empirically derived curves. Ann. Bot. 61, 127–138 [Google Scholar]
- 29. Wharton M. J. (1955) The use of tetrazolium test for determining the viability of seeds of the genus Brassica. Proc. Int. Seed Test. Assoc. 20, 81–88 [Google Scholar]
- 30. Berridge M. V., Tan A. S., McCoy K. D., Wang R. (1996) The biochemical and cellular basis of cell proliferation assays that use tetrazolium salts. Biochemica 4, 725–732 [Google Scholar]
- 31. Lindqvist Y., Schneider G. (1993) Thiamin diphosphate dependent enzymes: transketolase, pyruvate oxidase and pyruvate decarboxylase. Curr. Opin. Struct. Biol. 3, 896–901 [Google Scholar]
- 32. Machado C. R., de Oliveira R. L., Boiteux S., Praekelt U. M., Meacock P. A., Menck C. F. (1996) Thi1, a thiamine biosynthetic gene in Arabidopsis thaliana, complements bacterial defects in DNA repair. Plant Mol. Biol. 31, 585–593 [DOI] [PubMed] [Google Scholar]
- 33. Næsted H., Frandsen G. I., Jauh G. Y., Hernandez-Pinzon I., Nielsen H. B., Murphy D. J., Rogers J. C., Mundy J. (2000) Caleosins: Ca2+-binding proteins associated with lipid bodies. Plant Mol. Biol. 44, 463–476 [DOI] [PubMed] [Google Scholar]
- 34. Richard J. P. (1993) Mechanism for the formation of methylglyoxal from triosephosphates. Biochem. Soc. T. 21, 549–553 [DOI] [PubMed] [Google Scholar]
- 35. Ahmad A., Abdin M. Z. (2000) Effect of sulphur application on lipid, RNA and fatty acid content in developing seeds of rapeseed (Brassica campestris L.). Plant Sci. 150, 71–76 [Google Scholar]
- 36. Dixon D. P., Skipsey M., Edwards R. (2010) Roles for glutathione transferases in plant secondary metabolism. Phytochemistry 71, 338–350 [DOI] [PubMed] [Google Scholar]
- 37. Eastmond P. J., Graham I. A. (2001) Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci. 6, 72–78 [DOI] [PubMed] [Google Scholar]
- 38. Higgins T. J. V., Chandler P. M., Randall P. J., Spencer D., Beach L. R., Blagrove R. J., Kortt A. A., Inglis A. S. (1986) Gene structure, protein structure, and regulation of the synthesis of a sulfur-rich protein in pea seeds. J. Biol. Chem. 261, 11124–11130 [PubMed] [Google Scholar]
- 39. Hirai M. Y., Fujiwara T., Chino M., Naito S. (1995) Effects of sulfate concentrations on the expression of a soybean seed storage protein gene and its reversibility in transgenic Arabidopsis thaliana. Plant Cell Physiol. 36, 1331–1339 [PubMed] [Google Scholar]
- 40. Tabe L. M., Droux M. (2002) Limits to sulfur accumulation in transgenic lupin seeds expressing a foreign sulfur-rich protein. Plant Physiol. 128, 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu B., Gruber M., Khachatourians G., Hegedus D. D., Hannoufa A. (2010) Gene expression profiling of developing Brassica napus seed in relation to changes in major storage compounds. Plant Sci. 178, 381–389 [Google Scholar]
- 42. D'Hooghe P., Bataillé M.-P., Trouverie J., Avice J.-C. (2013) A specific method of 34S labelling provides evidence that sulphate assimilation occurs in developing seeds and pod walls of Brassica napus L. subjected to ample or limited S nutrition. Rapid Commun. Mass Spectrom. 27, 2737–2744 [DOI] [PubMed] [Google Scholar]
- 43. Andréasson E., Jørgensen L. B., Höglund A.-S., Rask L., Meijer J. (2001) Different myrosinase and idioblast distribution in Arabidopsis and Brassica napus. Plant Physiol. 127, 1750–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dubuis P. H., Marazzi C., Städler E., Mauch F. (2005) Sulphur deficiency causes a reduction in antimicrobial potential and leads to increased disease susceptibility of oilseed rape. J. Phytopathol. 153, 27–36 [Google Scholar]
- 45. Ahn I.-P., Kim S., Lee Y. (2005) Vitamin B1 functions as an activator of plant disease resistance. Plant Physiol. 138, 1505–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nikiforova V. J., Kopka J., Tolstikov V., Fiehn O., Hopkins L., Hawkesford M. J., Hesse H., Hoefgen R. (2005) Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol. 138, 304–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rajjou L., Duval M., Gallardo K., Catusse J., Bally J., Job C., Job D. (2012) Seed germination and vigor. Annu. Rev. Plant. Biol. 63, 507–533 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.