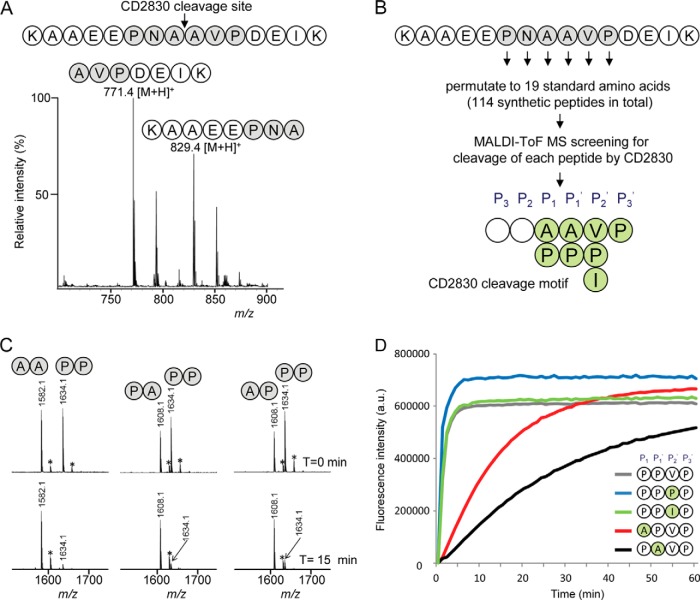
Fig. 3.
Identification of the CD2830 cleavage motif based on a peptide library screen reveals a preference for cleavage between proline residues. A, MALDI-TOF-MS spectrum of a synthetic peptide containing the HSP90β sequence AAEEPNAAVPDEI (amino acids 696–708) after 16 h of incubation with rCD2830. B, a synthetic peptide library was constructed in which all six positions surrounding the CD2830 scissile bond were permutated to the 19 standard amino acids within a core synthetic peptide (KAAEEPNAAVPDEIK), resulting in a total set of 114 peptides. Peptides were individually incubated with rCD2830, and cleavage was measured via MALDI-TOF-MS. The resulting CD2830 cleavage motif based on this peptide library screen is shown. C, binary mixtures of synthetic peptides with either a proline or an alanine at the P1 and P1′ positions in the core peptide (see above) were incubated with rCD2830 and analyzed via MALDI-TOF-MS after 15 min of incubation. After this short incubation, cleavage of the peptide containing a proline at both P1 and P1′ was more efficient than cleavage of the peptides containing one or two alanines. *+Na+. D, within a core synthetic FRET peptide (DabcylLys-EVNPPVPD-EdansGlu), permutations were introduced at the P1, P1′, and P2′ (PPV) positions. All peptides (50 μm) were incubated with rCD2830, and the formation of cleavage products was followed in time using fluorescence detection (see “Experimental Procedures” for details).