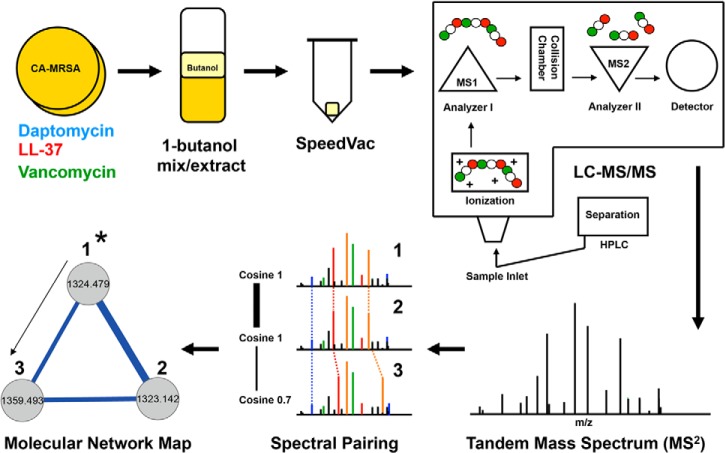
Fig. 1.
Mass spectrometry-based molecular networking. CA-MRSA USA 300 strain TCH1516 was grown to late stationary phase alone or in the presence of antibiotic agents (1). The organic based solvent 1-butanol was used to extract small molecules and peptides from the CA-MRSA growth culture (2). The 1-butanol extract was brought to complete dryness by the use of a SpeedVac centrifuge (3). The solid extract was re-suspended in H2O and then components were separated by HPLC and directly electrosprayed into a Thermo Finnegan LTQ (4) for tandem MS analysis (5). The resulting data sets were processed by Spectral Networks (6), which compiles and clusters similar overlapping spectra based on similarly scores expressed as cosine values. A graphical representation of the clustered spectra population was displayed by the use of Cytoscape network maps. Individual tandem mass spectra are represented as circular nodes that are connected to other nodes with similar fragmentation patterns (7). Nodes within subpopulation clusters can be targeted based on theoretical parent ion masses. Once a validated node is identified (denoted as 1* in schematic), the structural information can be used as a building block to propagate and identify adjacent nodes in the cluster.