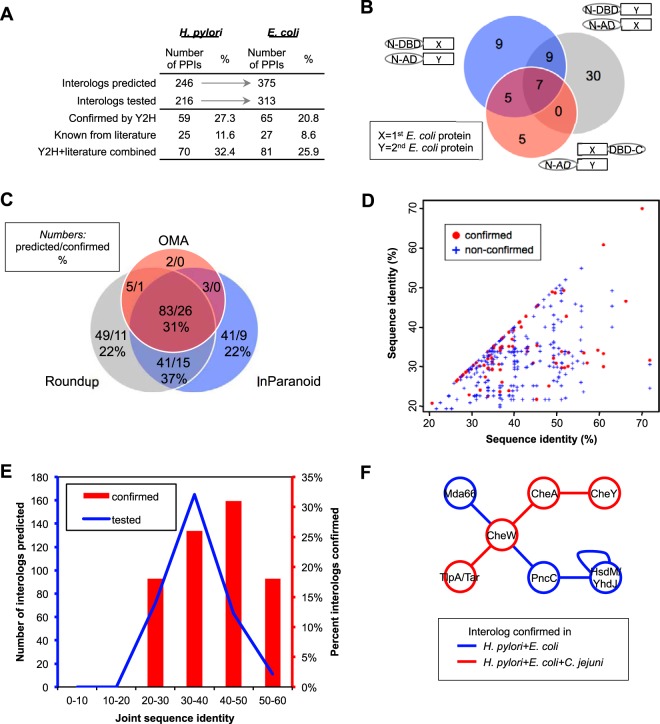
Fig. 5.
Conservation test of interologs and conserved subnetworks. A, Summary of conservation tests carried out in this study by pairwise Y2H assays using protein pairs of E. coli and H. pylori PIM.2_all as prediction source. In addition, interologs from PPI databases and the literature are compared. Detailed results are given in supplemental Table S5 and S6. B, Number of interactions tested as positive with E. coli pairs by different Y2H vectors. Note that both N- and C-terminal fusions were used as indicated, while H. pylori screens only used N-terminal fusions. C, Numbers of E. coli interologs predicted (first number) and verified (second number) by various prediction methods. The confirmation rate is given in percent. For details see supplemental Table S7. D, Protein pairs of the experimentally confirmed and nonconfirmed interactions from our pairwise E. coli Y2H tests and literature overlaps are plotted against their sequence identities (%). Each dot represents an interolog in E. coli and the axis represents the identity of each protein with its ortholog in H. pylori. Solid, red circles show positively verified interactions, blue crosses are not confirmed ones. See also supplemental Fig. S2. E, Comparison of the joint sequence identities versus predicted and confirmed interologs. See text for details. F, Example of a conserved subnetwork enriched for “signal transduction” and chemotaxis proteins (TlpA, CheW, CheA, CheY). “Red PPIs” were identified in H. pylori, E. coli, and C. jejuni and represent a part of the well understood chemotaxis pathway. “Blue PPIs” were found in H. pylori and E. coli, only, and involve the unexpected enzymes Mda66 (MdaB, quinone reductase), PncC (NMN amidohydrolase), and YhdJ (DNA adenine methyltransferase).