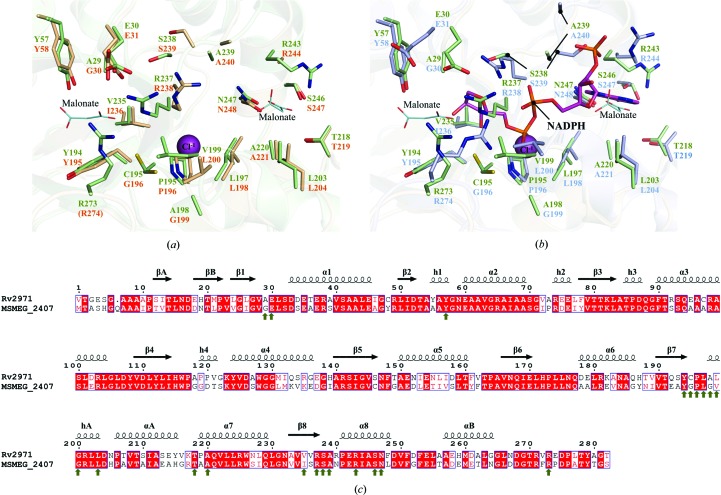
Figure 4.
Structural comparison between Rv2971 and MSMEG_2407. Ball-and-stick representation of the NADPH binding pockets of Rv2971 and (a) MSMEG_2407 in its apo form (PDB entry 2wzt) and (b) in its holoenzyme form in the presence of NADPH (PDB entry 2wzm). Visualized is the structural comparison between amino-acid residues in contact with NADPH in the MSMEG_2407 holoenzyme structure with residues of identical position in Rv2971. Residues of Rv2971, the MSMEG_2407 apo form and the MSMEG_2407 holoenzyme are represented in green, wheat and blue, respectively. Bound malonate ions and a single chloride ion are represented as cyan lines and magenta spheres, respectively. The bound NADPH molecule in the MSMEG_2407 holoenzyme is represented in pink. Missing residues are indicated in parentheses. (c) Sequence alignment between Rv2971 and MSMEG_2407. The alignment was prepared with the program ClustalW2 and visualized using ESPript v.2.2. The secondary-structure elements correspond to the structure of Rv2971. Strict sequence-identical residues are denoted with a red background, while similar residues are visualized in red text with white background. Sequence similarities in groups are denoted by blue boxes. MSMEG_2407 residues interacting with NADPH by hydrogen bonding or van der Waals interactions are indicated by green arrows.