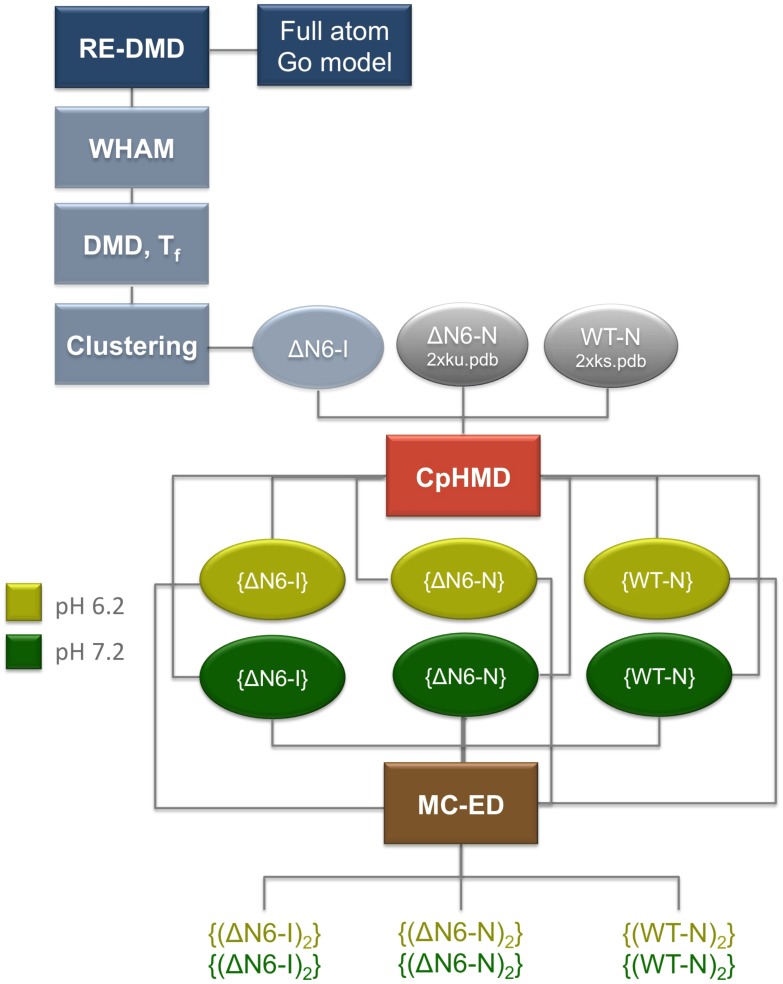
Figure 7. Methodological flow-chart.
The methodological approach used in this work is based on a three-stage process. In the first stage, equilibrium sampling is performed with a full atomistic Gō model combined with replica-exchange discrete molecular dynamics simulations (RE-DMD). The transition temperature is calculated as the temperature at which the heat capacity peaks, and the heat capacity is computed from the mean square fluctuations in energy at each temperature considered in the RE simulations. The calculation of the free energy surfaces at the transition temperature – which indicates the existence of an intermediate state for ΔN6 – is done with the Weighted Histogram Analysis Method (WHAM). Structural clustering is performed over ensembles of conformations extracted from DMD simulations at the transition temperature to isolate the intermediate state populated by ΔN6 that is termed ΔN6-I. In the second stage, constant-pH molecular dynamics simulations (CpHMD) are used to study the effects of pH on the structure of ΔN6-I, native structure of ΔN6 (ΔN6-N) and native structure of the wild-type Hβ2m (WT-N). Two pH values are investigated, namely pH 6.2 and pH 7.2. The output of the CpHMD simulations consists of six ensembles of equilibrated conformations of ΔN6-I, ΔN6-N and WT-N at the two considered pH values. Finally, in the last stage of our approach, the conformations extracted from the CpHMD simulations are used as input for the Monte Carlo Ensemble Docking (MC-ES), which delivers ensembles of dimers of ΔN6-I, ΔN6-N and WT-N at the two considered pH values. The study of the dimerization interface is based on the statistical analysis of the ensembles of dimers.