Figure 4. Human NP exhibits increased evolution in CTL epitopes relative to swine NP.
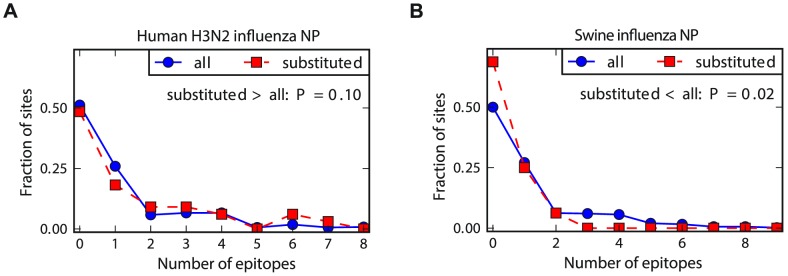
The number of CTL epitopes per site for all sites in NP versus those that substituted along the evolutionary trajectories for (A) human and (B) swine influenza. In human influenza, the substituted sites contain more epitopes than average sites – but in swine influenza, the substituted sites contribute to fewer epitopes than average sites. The P-values on the plots are the fraction of random subsets of all sites that contain as many (human NP) or as few (swine NP) total epitopes as the sites that actually substituted during the natural evolution of that homolog. The hypothesis of greatest interest is whether the substituted sites in the human NP contain more epitopes than do substituted sites in the swine NP. To test this hypothesis, we drew paired random subsets of sites from the human and swine NP homolog of the same size as the actual numbers of substituted sites for each homolog, and determined the fraction of these paired random subsets in which the number of epitopes for the human NP exceeded that for the swine NP by at least as much as for the actual data. This test gives a P-value of 0.008, supporting the hypothesis that human NP exhibits an increased rate of evolution in epitopes relative to swine NP. See http://jbloom.github.io/epitopefinder/example_NP_CTL_epitopes_H3N2_and_swine.html for code, input data, and detailed documentation.
