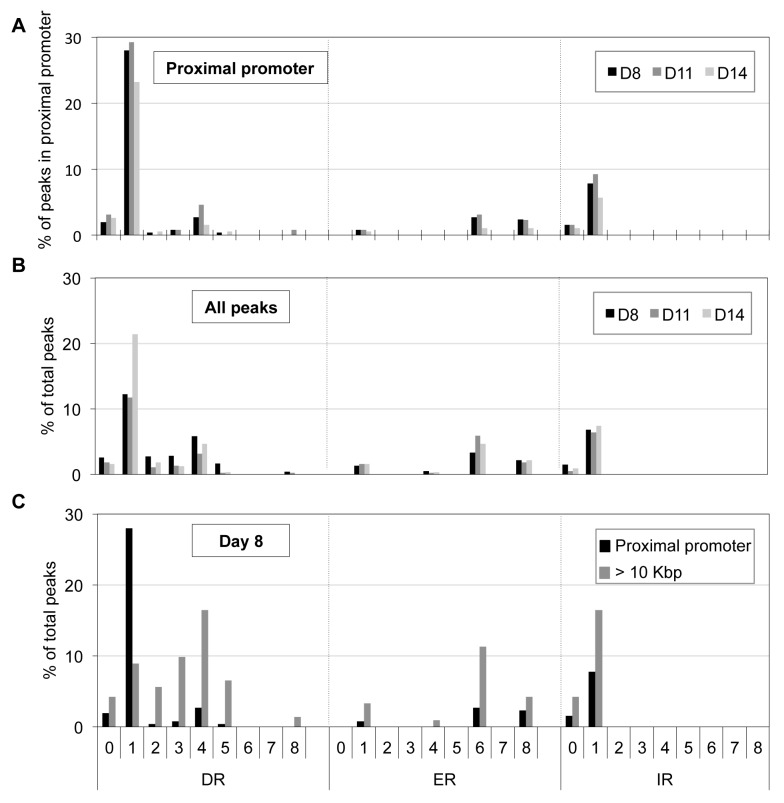
Figure 5. Distribution of potential NR binding sites in TR4-bound peaks.
Peak sequences (250 bp from peak center) were interrogated using NHR-scan for the presence of direct repeat (DR), everted repeat (ER) and inverted repeat (IR) motifs with 0–8 spacer nucleotides in D8, D11 and D14 (A, B) or in D8 (C) cells. The percentages of each motif type in peaks located at the proximal promoter (A), in all peaks (B), and in peaks at proximal promoter vs. >10 Kbp from genes (C) are represented graphically.