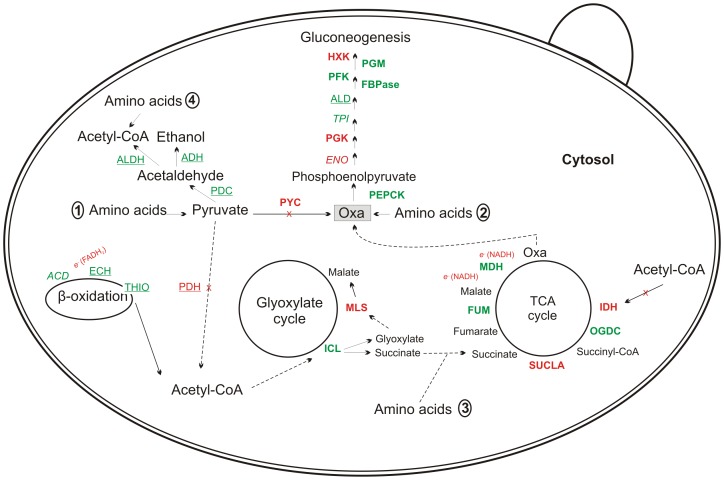
Figure 3. Overview of metabolic responses of Paracoccidioides to carbon starvation.
The figure summarizes the data from transcriptome and proteomic analyses and suggests the mechanism and the first flow of carbon used by this fungus to overcome the carbon starvation stress. HXK: hexokinase; PGM: phosphoglucomutase; PFK: 6-phosphofructokinase; FBPase: fructose-1,6-biphosphatase; ALD: fructose-bisphosphate aldolase; TPI: triosephosphate isomerase; PGK: phosphoglycerate kinase; ENO: enolase; PEPCK: phosphoenolpyruvate carboxykinase; PYC: pyruvate carboxylase; PDC: pyruvate decarboxylase; ADH: alcohol dehydrogenase; ALDH: aldehyde dehydrogenase; ACD: acyl-CoA dehydrogenase; ECH: enoyl-CoA hydratase; THIO: 3-ketoacyl-CoA thiolase; PDH: pyruvate dehydrogenase; ICL: isocitrate lyase; MLS: malate synthase; IDH: isocitrate dehydrogenase; OGDC: 2-oxoglutarate dehydrogenase E1; SUCLA: succinyl-CoA ligase; FUM: fumarate hydratase (fumarase) and MDH: malate dehydrogenase. Enzymes were colored according to their differences in expression and labeled to indicate whether the data were obtained from transcriptome or proteomics. Italic, bold, and underlined labels indicate that the data were obtained from transcriptome, proteome, or both, respectively. Green or red indicate up- or down-regulated proteins, respectively. The numbers 1, 2, 3, and 4 indicate up-regulated amino acids involved in pyruvate, oxaloacetate, succinate, and acetyl-CoA production, respectively. 1) pyruvate production: tryptophan and cysteine. 2) oxaloacetate production: phenylalanine, glutamate and tyrosine. 3) succinate production: threonine. 4) acetyl-CoA production: threonine, tryptophan, tyrosine and leucine. Italic, bold, and underlined labels indicate that the amino acids accumulations were obtained from transcriptome, proteome, or both analyzes, respectively. OXA: oxaloacetate; e−: released electrons from enzymatic reaction.