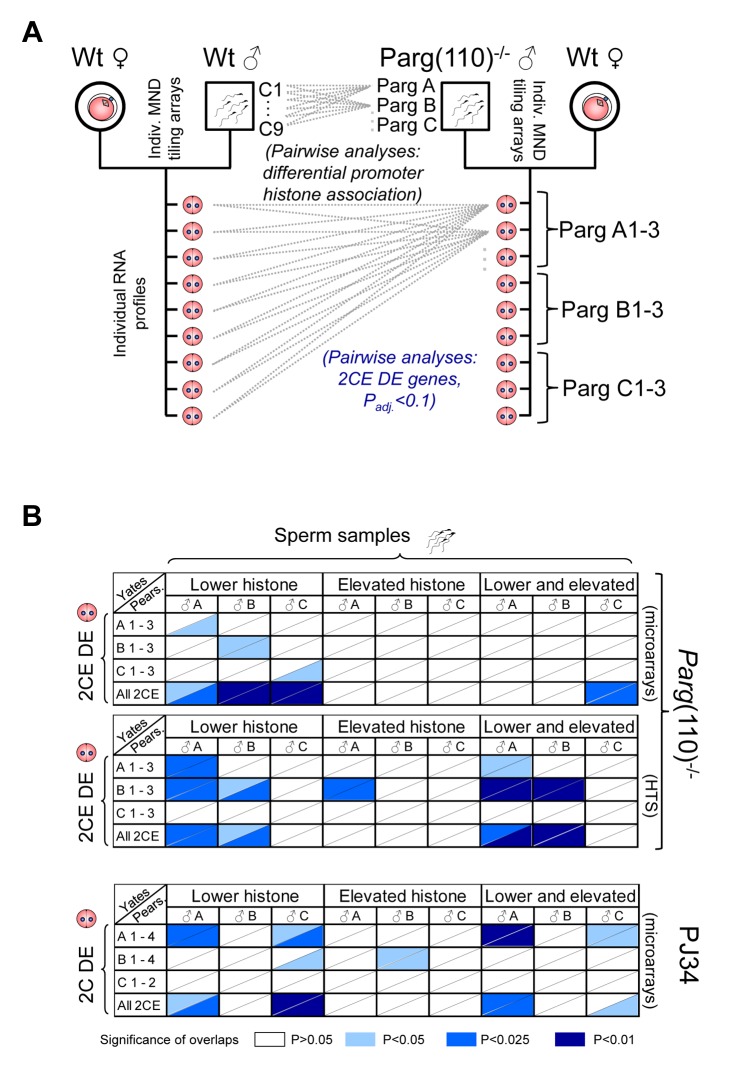
Figure 4. Differential sperm histone association of genes in Parg(110)−/− and PJ34-treated males is significantly associated with differential expression of these genes in offspring 2-cell embryos.
(A) Outline of the comparison procedure, shown here only for the Parg(110) gene disrupted mouse model. A similar regimen was used for the PJ34-treated males and their offspring. Differential histone association of genes in sperm from individual males (PargA, PargB, PargC) was determined by pair-wise comparison with all individual wild-type data sets obtained from 9 individually analyzed control males. Differential gene expression was determined by pair-wise comparison of individual offspring from the Parg(110)−/− males (PargA1-PargC3) with all 9 individual wild-type 2-cell embryo data sets by either microarrays or next generation sequencing. Differential gene expression was determined using ANOVA analyses and adjusted P-value calculation (with Padj<0.1 considered to be significant). (B) Pearson (uncorrected) and Yates (corrected) Chi-squared tests were used to determine the significance of overlaps of the lists of genes that were differentially histone associated in sperm samples of the sires (“Sperm samples”) compared to controls (Parg A–C and PJ34A–C), with the lists of genes that were differentially expressed in at least one of the 3 or 4 offspring 2-cell embryos from these sires (“2CE DE”, Parg(110)−/−: A 1–3, B 1–3, C 1–3, and for PJ34: A1–4, B1–4, C1–2). A genetic background of 19,472 genes interrogated by the microarrays and 20,018 genes interrogated by the tiling arrays and sequencing platforms was used for the calculations. Ranges of P-values resulting from Yates or Pearson are indicated in different shades of blue if P≤0.05, i.e., the overlaps were significant (see color legend in figure). The P-value denotes the confidence with which the null-hypothesis can be dismissed that the overlap between the list of genes with abnormal histone association in the sire with the list of genes that are DE in the offspring could be predicted by statistical probability, i.e. coincidence. Upper two panels: Parg(110)−/− group of fathers and offspring embryos, microarray expression analyses with either Yates' chi-squared test (upper triangular portion of each cell) or Pearson's (lower triangular portion of cells). HTS: Parg(110) group, high throughput sequencing of 2CE gene expression. Lower panel: overlaps of PJ34-treated group of fathers (sperm) and offspring embryos (2-cell embryo differential gene expression, “2CE DE”). Note that mainly overlaps between genes with lower histone retention in sperm and differentially expressed genes in offspring embryos are significant with P≤0.05 (left 1/3 portion of each panel).