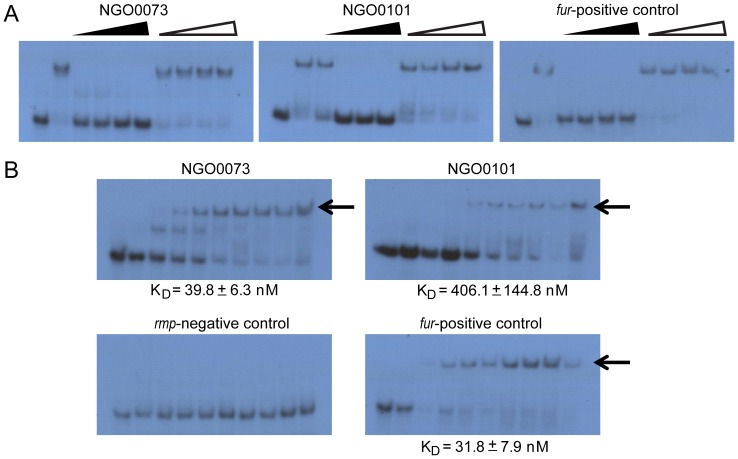
Figure 2. Fur binding to the predicted Fur boxes. (A) Cold competition assay for the specificity of Fur binding to the predicted Fur boxes.
32P-labeled ∼50 bp dsDNAs were analyzed after incubation with Fur and unlabeled dsDNA probes (cold probes). Two types of cold competitor probes were used: fur, which contains a Fur box, and rmp, which does not bind to Fur. When the unlabeled probes fur compete out binding of labeled probes, but the unlabeled rmp cannot, the binding of Fur to the predicted Fur box was considered specific. Lane 1, free 32P-labeled DNA; Lane 2 through Lane 10 contained gonococcal Fur. For the fur and NGO0073 Fur box, Fur protein was added at a concentration of 100 nM and for the NGO0101 Fur box, 400 nM of Fur protein was added. The fold excess of the cold probes was increased from Lane 3 to Lane 6 and Lane 7 to Lane 10 ranging from 50 fold, 500 fold to 1000 fold (indicated by the triangles). (B) Fur binding affinities determined by EMSA. The 32P labeled dsDNA probes were incubated with a gradient of concentrations of purified gonococcal Fur protein (Lane 1 to Lane 10, 0 nM, 5 nM, 25 nM, 50 nM, 100 nM, 200 nM, 400 nM, 600 nM, 800 nM and 1000 nM, respectively). Arrows indicate the shift of Fur-bound probes. The apparent binding affinities (KD) were calculated using GraphPad Prism and were represented as mean ± standard error. The gene designations of N. gonorrhoeae F62 were assigned according to their homologues in N. gonorrhoeae FA1090.