Abstract
It is still challenging to prevent and treat respiratory infectious diseases. One critical step in the successful treatment of respiratory infections is rapid diagnosis by identifying the causative microorganisms in a timely fashion. However, traditional methods for identification of causative agents could not satisfy the need for rapid and accurate testing due to the limitations of technology-used. In recent years, matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS) has been validated and used for rapid identification of microorganism and for potential discovery of diseases associated biomarkers. We reviewed recent advances of MALDI-TOF-MS as the laboratory diagnostic tool for the rapid laboratory diagnosis of microorganisms associated with respiratory infectious diseases, with the focus on rapid identification of pathogenic bacteria and molecular markers discovery using MALDI-TOF-MS. With the advanced technologies such as MALDI-TOF, early and targeted therapies based on rapid identification of pathogens and could lead to quick and effective treatment of respiratory infections and better patient management.
Keywords: Respiratory infectious diseases, rapid laboratory diagnosis, matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS), pathogenic bacteria
Introduction
According to a recent report offered by World Health Organization (WHO), respiratory infections are the third leading cause of death worldwide and result in over three million deaths per year (1). Community acquired and hospital acquired respiratory infections are common and can be serious which can lead to complications such as pneumonia, bronchitis, sinus infections, and a general worsening of chronic conditions (2). As is the case with other infectious diseases, management of respiratory infections often requires a definitive laboratory identification of the causative agents in addition to clinical diagnosis for quick and direct optimal treatment.
Laboratory diagnosis of pathogens causing respiratory infections depends on many factors, which include but not limit to pathogen types and pathogen loads, specimen types, laboratory tools and methods used for staining, culture and detection.
The conventional microbial diagnosis of pathogens primarily relies on the culture of respiratory specimens such as sputum. Other respiratory specimen types include induced sputum, tracheal aspiration, bronchial wash, bronchial brush, and bronchoalveolar lavage (BAL). The quantification of colony forming unit (CFU) of certain specimens can be performed. If microorganism were recovered from culture, antimicrobial susceptibility testing (AST) should be subsequently performed to determine the antibiotic resistance (3). Unfortunately, traditional methods for microorganism identification take a few days and could not satisfy the need for rapid testing. Thus, rapid laboratory diagnosis of respiratory infections has been challenging.
Recently, the technology called matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) has been demonstrated in identifying microorganisms at species level. MALDI-TOF MS is regarded as a novel rapid clinical diagnostic strategy for identification of pathogens infections (4). Here, we review recent advances of MALDI-TOF-MS as the laboratory diagnostic tools for the rapid diagnosis of microorganisms associated with infectious diseases, and impact on AST and study.
Common pathogenic microorganisms and conventional diagnostic methods
There are many types of pathogenic microorganisms that can cause respiratory infections. Among those pathogens, Streptococcus pneumonia, Hemophilus influenza and Moraxella catarrhalis are most common in causing bacterial respiratory tract infections including acute otitis media (AOM), sinusitis, laryngitis and pneumonia. In addition, Legionella pneumophila is mainly responsible for atypical pneumonia, and Mycobacterium tuberculosis leads to tuberculosis, etc. However, the etiologic agent remains unknown in nearly 30% of cases, suggesting that the existing methods lack accuracy and sensitivity for identification of unknown pathogens including viral pathogens in respiratory infection cases.
Gram stain and culture of respiratory samples have been traditionally used for the detection and distinguish of bacterial pathogens. In general, it takes 48 to 96 hours to have bacterial identification and resistance determination. In the meantime when identification and resistant results are not available, patients may be treated with broad-based antibiotics that are often unnecessary, inappropriate, even harmful, which eventually lead to resistance, even multiple resistances. Thus, new technologies are needed to shorten the time of detection, identification, and AST and to provide rapid laboratory results to clinicians for better patient care and control of antimicrobial resistance.
MALDI-TOF mass spectrometry (MALDI-TOF-MS), a new diagnostic technology for rapid diagnosis of pathogenic microorganisms in respiratory infection
Rapid identification of pathogenic bacteria using MALDI-TOF-MS
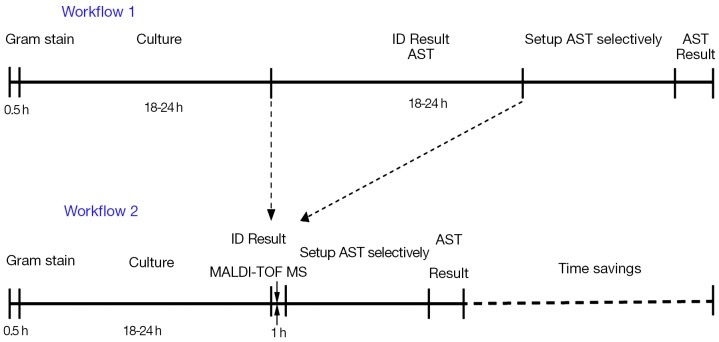
In general, earlier diagnosis of respiratory infections can lead to better outcomes. Generally, MALDI-TOF-MS provides effective bacterial identification and antibiotic resistance determination directly from positive blood culture specimens up to 24 hours faster than conventional method (Figure 1). Targeted therapies, based on the rapid identification of causative pathogens and antimicrobial resistance, shall become more effective and timely. It is essential for each clinical microbiology laboratory to develop techniques that identify these bacteria correctly and rapidly. Advanced microbiological methods are important for earlier bacteria detection. MALDI-TOF-MS has shown its fast and reliable feature for identifying microorganisms including pathogenic bacteria and yeasts from samples with positive culture, which may represent a rapid, inexpensive and alternative assay for identification of bacteria at the species level.
Figure 1.
Comparison of new method workflow with traditional culture workflow. In the ideal situation when there is one predominant colony grow from the culture. If there are mixed cultures with different colonies, the time of saving could be two days as the subcultures are needed for pure colony and for identification by conventional method. Workflow 1, conventional culture; workflow 2, new method with MALDI-TOF MS; ID, identification; AST, antimicrobial susceptibility testing.
Here we only list a few studies to prove the concept. The Streptococcus mitis group is a set of closely related species in which conventional identification methods cannot reliably make differentiation analysis. The most important pathogen within the S. mitis group, S. pneumonia, is conventionally distinguished from the others on the basis of its susceptibility to optochin or its solubility in bile (5). In a recent study, the Vitek MS v2.0 System (MALDI-TOF-MS technology) accurately distinguished Streptococcus pneumonia from nonpneumococcal S. mitis group species. Only 1 of 116 nonpneumococcal isolates (<1%) was misidentified as S. pneumoniae. None of 95 pneumococcal isolates was misidentified. In this case, MALDI-TOF-MS provides a rapid, simple means of discriminating among these challenging organisms (6).
Infections caused by Legionella species other than L. pneumophila are also often lack of laboratory diagnosis, owing to limitations of the conventional diagnostic methods that are biased towards the detection of L. pneumophila (7). Recently, MALDI-TOF-MS was used to identify the specific species of the Legionella non-pneumophila isolates; and the result was consistent with the data of reference method and macrophage infectivity potentiator gene (mip) sequencing (8).
Corynebacteria have been recognized as opportunistic pathogens in causing various types of healthcare-associated infections in immunocompromised hosts (9). Identification of Corynebacteria by conventional methods is suboptimal and it is likely that their true prevalence in clinical specimens either as colonizers or as pathogens remains largely underestimated. Rapid identification of toxigenic Corynebacterium species and non-diphtheria Corynebacterium, including C. triatum can be achieved by using the MALDI-TOF MS (10).
Another application of MALDI TOF is the identification of yeasts. As the number of patients with profound immunosuppression continues to rise, the morbidity and mortality burdens due to invasive fungal infections are significant. Traditional methods used to identify clinical yeast isolates are time-consuming and may result in low-discrimination identifications (11). In a recent study MALDI-TOF Vitek MS was applied on the identification of yeasts isolated from clinical specimens. A collection of 852 isolates was tested, in total, 823 isolates (96.6%) were identified to the genus level and 819 isolates (96.1%) were identified to species level, 24 isolates (2.8%) were not identified and 5 misidentified, which indicated that MALDI-TOF-MS offers a balance between speed and highly accurate yeast identification (12).
MALDI-TOF-MS: our experience
Here at Grady Memorial Hospital in metropolitan Atlanta area, we believe the need of culture based methods for laboratory diagnosis of infection causing microorganisms. We started to see the emergence of community-associated methicillin-resistant Staphylococcus aureus (MRSA) USA300 genotype as a major cause of healthcare-associated blood stream infections as early as 2004 (13), and we noticed emergence of community-acquired MRSA USA300 Clone as the predominant cause of S. aureus skin and soft tissue Infections in Atlanta (14). The bacteria isolated from clinical cultures can be used for surveillance study of antimicrobial resistance (15) as well as the immune response study of antibiotic-resistant nosocomial bacteria (16). Study on current or new CLSI breakpoints of antimicrobial susceptibility (17) and even the population-based active surveillance method for multidrug resistant gram-negative bacilli (18) or the inoculum effect study among bloodstream isolates of methicillin-susceptible Staphylococcus aureus (19) rely on positive isolates from cultures.
For the above reasons, we recognized the importance of clinical utility of new culture based technologies such as MALDI-TOF MS as an important laboratory tool to battle antibiotic resistance (20) and we potential tool for improved clinical microbiology laboratory automation (21).
We reported a few clinical cases as evidence-based medicine for clinical utility of MALDI-TOF MS to help diagnosis of rare case of bacterial infection (22) and rare case of fungal infection (23). We tried to improve the method for rapid identification of bacteria and yeasts from positive blood culture bottles by using a lysis-filtration method and MALDI-TOF mass spectrum analysis (24), and recently we had success in making the same day identification and full panel AST of bacteria from positive blood culture bottles possible by a combined lysis-filtration method with MALDI-TOF VITEK mass spectrometry and the VITEK2 system (25). Those methods can certainly help rapid laboratory diagnosis of respiratory infections.
In addition to bacterial and yeast identification, we studied the method to improve the diagnosis of mycobacteria including M. tuberculosis, an important respiratory pathogen. We compared the heat inactivation method and cell disruption protocols for identification of mycobacteria from solid culture media using MALDI-TOF VITEK MS (26).
Translation research: molecular markers discovery in microorganisms by MALDI-TOF MS
MALDI-TOF MS can be used for translational study. Most proteins in bacteria are adhesins/virulence factors that not only can trigger the immune response, but also aid bacteria to evade the host defense. Identification these proteins shall be helpful to understand pathogenic mechanisms and resistance mechanisms so that clinicians can optimize the treatment plan to ensure the correct and timely therapy.
Outer membrane vesicles (OMVs) secreted by bacteria can be recognized as long-distance delivery vehicles which transport diverse virulence factors and allow pathogens to interact with the host. A total of 57 proteins in Moraxella Catarrhalis OMVs were identified by using MALDI-TOF-MS analysis and gained new insights in the biological function of these protein-carrying lipid structures (27), and a novel molecular basis could play a role of OMVs in Moraxella Catarrhalis pathogenesis (27).
Surface enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS) can be considered as an extension of MALDI-TOF MS, which utilizes ProteinChip arrays with a MALDI-TOF-MS-based analytical platform, has been used for the discovery of diseases associated biomarkers (28). Tuberculosis is a major global concerned disease; its diagnosis is always problematic especially for latent tuberculosis and early disease. Recently, two biomarker-proteins, transthyretin and SAA des Arg were detected by SELDI-TOF-MS, which combined with neopterin and C-reactive protein, significantly improved the sensitivity and specificity for tuberculosis diagnosis (>80%) (29). Thus, MALDI-TOF-MS and its extension SELDI-TOF-MS shows promise in identifying novel biomarkers in respiratory infection that lead to advances in our understanding in pathophysiology and is useful for monitoring disease progression or treatment.
Summary
Microbiological diagnosis is often required to confirm clinical suspicions concerning infections. It is of great importance that microbiological diagnostic tools become more efficient and powerful, particularly for polymicrobial infections. MALDI-TOF MS as a new frontier technology is likely to have a major impact in clinical microbiology and provide platform for microbiology laboratory automation. The clinical utility of culture and proteomic based MALDI-TOF MS technologies by the microbial laboratories will enable laboratory diagnosis of common and rare pathogenic agents for respiratory infections quickly and accurately. More studies on identification of mycobacterial and fungal infection and clinical outcome analysis are needed.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- 1.The 10 leading causes of death in the world, 2000 and 2011. Available online: http://www.who.int/mediacentre/factsheets/fs310/en/
- 2.Bousbia S, Raoult D, La Scola B.Pneumonia pathogen detection and microbial interactions in polymicrobial episodes. Future Microbiol 2013;8:633-60 [DOI] [PubMed] [Google Scholar]
- 3.Torres A, El-Ebiary M.Bronchoscopic BAL in the diagnosis of ventilator-associated pneumonia. Chest 2000;117:198S-202S [DOI] [PubMed] [Google Scholar]
- 4.Bizzini A, Greub G.Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin Microbiol Infect 2010;16:1614-9 [DOI] [PubMed] [Google Scholar]
- 5.Wessels E, Schelfaut JJ, Bernards AT, et al. Evaluation of several biochemical and molecular techniques for identification of Streptococcus pneumoniae and Streptococcus pseudopneumoniae and their detection in respiratory samples. J Clin Microbiol 2012;50:1171-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branda JA, Markham RP, Garner CD, et al. Performance of the Vitek MS v2.0 system in distinguishing Streptococcus pneumoniae from nonpneumococcal species of the Streptococcus mitis group. J Clin Microbiol 2013;51:3079-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newton HJ, Ang DK, van Driel IR, et al. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev 2010;23:274-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Svarrer CW, Uldum SA. The occurrence of Legionella species other than Legionella pneumophila in clinical and environmental samples in Denmark identified by mip gene sequencing and matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Microbiol Infect 2012;18:1004-9 [DOI] [PubMed] [Google Scholar]
- 9.Camello TC, Souza MC, Martins CA, et al. Corynebacterium pseudodiphtheriticum isolated from relevant clinical sites of infection: a human pathogen overlooked in emerging countries. Lett Appl Microbiol 2009;48:458-64 [DOI] [PubMed] [Google Scholar]
- 10.Díez-Aguilar M, Ruiz-Garbajosa P, Fernández-Olmos A, et al. Non-diphtheriae Corynebacterium species: an emerging respiratory pathogen. Eur J Clin Microbiol Infect Dis 2013;32:769-72 [DOI] [PubMed] [Google Scholar]
- 11.Sanguinetti M, Porta R, Sali M, et al. Evaluation of VITEK 2 and RapID yeast plus systems for yeast species identification: experience at a large clinical microbiology laboratory. J Clin Microbiol 2007;45:1343-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westblade LF, Jennemann R, Branda JA, et al. Multicenter study evaluating the Vitek MS system for identification of medically important yeasts. J Clin Microbiol 2013;51:2267-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seybold U, Kourbatova EV, Johnson JG, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin Infect Dis 2006;42:647-56 [DOI] [PubMed] [Google Scholar]
- 14.King MD, Humphrey BJ, Wang YF, et al. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann Intern Med 2006;144:309-17 [DOI] [PubMed] [Google Scholar]
- 15.Wang YF, Dowzicky MJ. In vitro activity of tigecycline and comparators on Acinetobacter spp. isolates collected from patients with bacteremia and MIC change during the Tigecycline Evaluation and Surveillance Trial, 2004 to 2008. Diagn Microbiol Infect Dis 2010;68:73-9 [DOI] [PubMed] [Google Scholar]
- 16.Band VI, Ibegbu C, Kaur SP, et al. Induction of human plasmablasts during infection with antibiotic-resistant nosocomial bacteria. J Antimicrob Chemother. 2014 doi: 10.1093/jac/dku047. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang P, Hu F, Xiong Z, et al. Susceptibility of extended-spectrum-beta-lactamase-producing Enterobacteriaceae according to the new CLSI breakpoints. J Clin Microbiol 2011;49:3127-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reno J, Schenck C, Scott J, et al. Querying automated antibiotic susceptibility testing instruments: a novel population-based active surveillance method for multidrug-resistant gram-negative bacilli. Infect Control Hosp Epidemiol 2014;35:336-41 [DOI] [PubMed] [Google Scholar]
- 19.Livorsi DJ, Crispell E, Satola SW, et al. Prevalence of blaZ gene types and the inoculum effect with cefazolin among bloodstream isolates of methicillin-susceptible Staphylococcus aureus. Antimicrob Agents Chemother 2012;56:4474-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang YF. The lab’s increasing arsenal of tools to battle antibiotic resistance. MLO Med Lab Obs 2011;43:41. [PubMed] [Google Scholar]
- 21.Wang YF. Clinical microbiology laboratory automation and laboratory diagnosis of infectious diseases: present and future. Chinese J Laboratory Medicine 2013;36:592-4 [Google Scholar]
- 22.Hayek SS, Abd TT, Cribbs SK, et al. Rare Elizabethkingia meningosepticum meningitis case in an immunocompetent adult. Emerg Microbes Infect 2013;2:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan AW, Cartwright EJ, Reddy SC, et al. Pichia anomala (Candida pelliculosa) fungemia in a patient with sickle cell disease. Mycopathologia 2013;176:273-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fothergill A, Kasinathan V, Hyman J, et al. Rapid identification of bacteria and yeasts from positive-blood-culture bottles by using a lysis-filtration method and matrix-assisted laser desorption ionization-time of flight mass spectrum analysis with the SARAMIS database. J Clin Microbiol 2013;51:805-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Machen A, Drake T, Wang YF. Same day identification and full panel antimicrobial susceptibility testing of bacteria from positive blood culture bottles made possible by a combined lysis-filtration method with MALDI-TOF VITEK mass spectrometry and the VITEK2 system. PLoS One 2014;9:e87870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machen A, Kobayashi M, Connelly MR, et al. Comparison of heat inactivation and cell disruption protocols for identification of mycobacteria from solid culture media by use of vitek matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 2013;51:4226-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaar V, de Vries SP, Perez Vidakovics ML, et al. Multicomponent Moraxella catarrhalis outer membrane vesicles induce an inflammatory response and are internalized by human epithelial cells. Cell Microbiol 2011;13:432-49 [DOI] [PubMed] [Google Scholar]
- 28.Vorderwülbecke S, Cleverley S, Weinberger SR, et al. Protein quantification by the SELDI-TOF-MS-based ProteinChip® System. Nat Methods 2005;2:393-5 [Google Scholar]
- 29.Shah HN, Gharbia SE. eds. Mass spectrometry for microbial proteomics. Chichester: John Wiley & Sons Ltd, 2010:244. [Google Scholar]