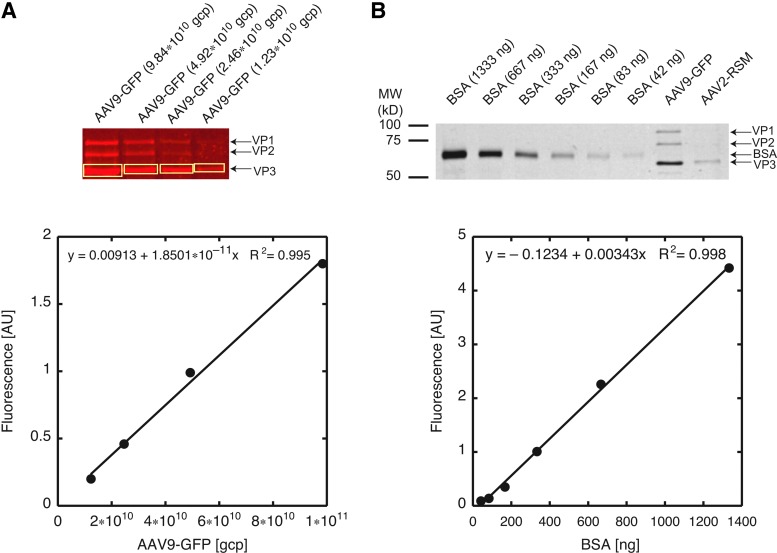
FIG. 1.
Quantification of viral particle titers by Coomassie Brilliant Blue-stained SDS–PAGE and infrared-fluorescence scanning with an Odyssey scanner. (A) The fluorescence intensity is linear over a wide range of viral particles loaded onto the gel. Top: The indicated amount of AAV9-GFP particles was separated on a 12%, reducing SDS–polyacrylamide gel, stained with Coomassie Brilliant Blue, followed by scanning of the wet gel with an Odyssey (LI-COR) scanner, using the 700-nm channel. v, viral particles. Brightness was adjusted uniformly across the entire image. Positions of the viral capsid proteins (VP1, VP2, and VP3) are indicated with arrows. Bottom: The number of viral particles loaded in each lane is plotted against integrated fluorescence of the boxed VP3 bands (top). Quantification was performed before adjusting the brightness. Regression analysis demonstrates the linearity of the integrated fluorescence intensity over the entire particle range covered. (B) The fluorescence intensity is linear over a wide range of BSA loaded onto the gel. Top: The indicated amounts of BSA were separated on a 12%, reducing SDS–polyacrylamide gel, stained with Coomassie Brilliant Blue, followed by scanning of the wet gel with an Odyssey (LI-COR) scanner, using the 700-nm channel. The picture was converted to grayscale and inverted. Brightness was adjusted uniformly across the entire image. Bottom: Quantification of the gel (top) demonstrates the linearity of the integrated fluorescence intensity over the entire range of BSA loaded. Regression analysis of the BSA standard curve allowed determination of the number of viral particles loaded in the two right-most lanes: AAV9-GFP (1.32×1011 viral particles) and AAV2-RSS (2.41×1010 viral particles). Positions of the viral capsid proteins (VP1, VP2, and VP3) and BSA are indicated with arrows, and the position and size of the molecular weight standards are shown on the left.