Figure 1.
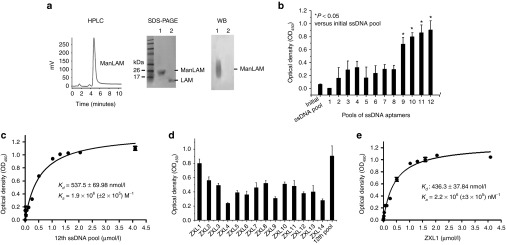
High-affinity aptamers for mannose-capped lipoarabinomannan (ManLAM) were generated by systematic evolution of ligands by exponential enrichment (SELEX). (a) Identification of the purified ManLAM from Mycobacterium tuberculosis (M. tb) H37Rv by high-performance liquid chromatography (HPLC) showing the final preparation of ManLAM, sodium dodecyl sulfate–polyacrylamide gel electrophoresis (silver staining), and western blot (WB) analyses. (b) After 12 rounds of screening against ManLAM, single-stranded DNA (ssDNA) pools from each round were analyzed for their binding to ManLAM by enzyme-linked oligonucleotide assay. Aptamer pools (2 µmol/l) were incubated in wells coated with ManLAM (40 µg/ml in phosphate-buffered saline). In the background control, 40 µg/ml of ManLAM was coated on the wells, but none of the ssDNA aptamers were added. For each sample, the optical density at 450 nm (OD450) of the background control was subtracted from the OD450 of experimental sample.*P < 0.05 versus initial ssDNA pool. All data are shown as the means ± SEMs (n = 3). (c) Binding of the 12th ssDNA pool to ManLAM. The 12th-round pool of ssDNA was incubated in wells coated with ManLAM. All data are shown as the means ± SEMs (n = 3). The Kd of 537.5 ± 69.98 nmol/l was determined as described in the Methods section. (d) Binding of the single aptamers ZXL1–ZXL14 to ManLAM. The single aptamers ZXL1–ZXL14 (2 µmol/l) were, respectively, incubated in wells coated with ManLAM. All data are shown as the means ± SEMs (n = 3). (e) Analysis of binding of ZXL1 to ManLAM. ZXL1 was added and incubated in the wells coated with ManLAM. All data are shown as the means ± SEMs (n = 3). The Kd of 436.3 ± 37.84 nmol/l was established as described in the Methods section.
