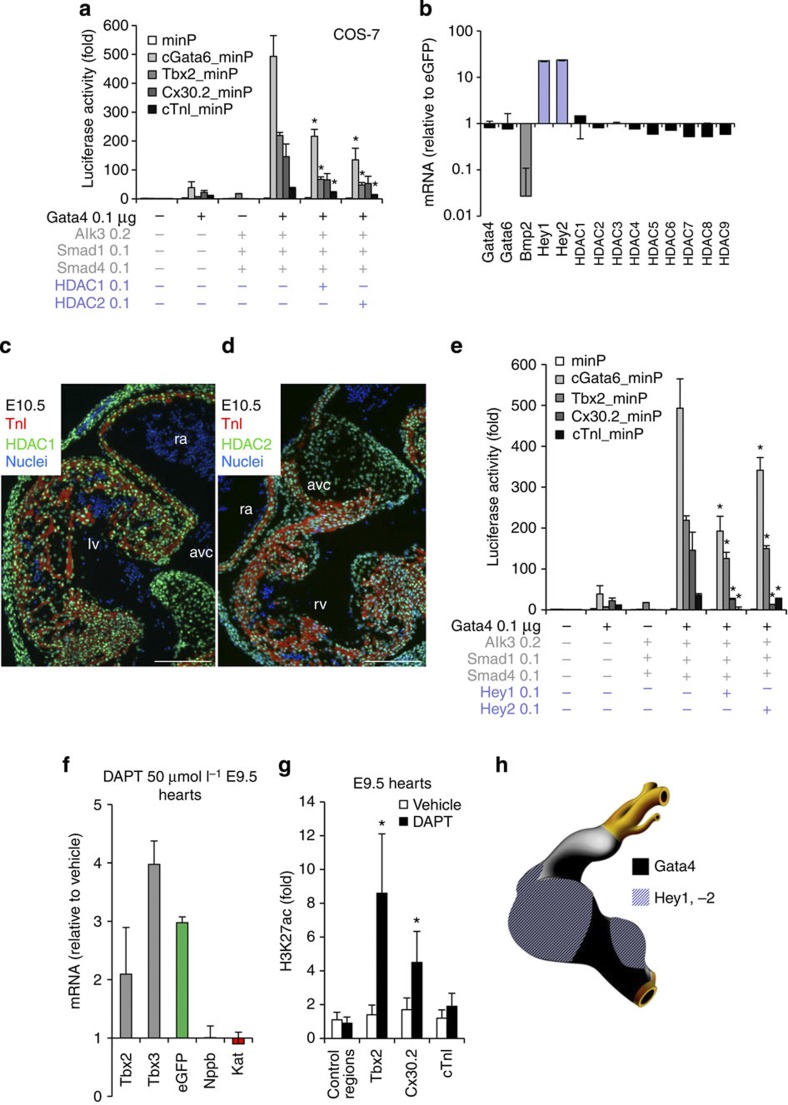
Figure 4. The Gata4/Hey/Hdac complex induces H3K27 deacetylation and AV canal gene repression.
(a) Luciferase reporter assays on 102 bp cGata6, the 380 bp Tbx2, 660 bp Cx30.2 and the 356 bp cTnI genomic fragments. Constructs were co-transfected with Gata4, Alk3CA, Smad1 and Smad4, HDAC1 and 2 expression vectors into Cos-7 cells (mean±s.e.m., n=6, *P<0.05 for Gata4, BMP2 effectors and HDAC1, 2 vs Gata4 and BMP2 co-transfection using two- and three-way analysis of variance (ANOVA)). (b) qPCR analysis of mRNA isolated from AV canal-EGFP and chambers–Katushka sorted cells. Values are normalized against EGFP population. Immunohistochemistry for Hdac1 (c) and 2 (d) proteins on sections of E10.5 hearts. Scale bars represent 100 μm. (e) Luciferase reporter assays on AV canal enhancers with expressing vectors encoding for Gata4, Bmp2 effectors and the notch target genes Hey1 and 2 (mean±s.e.m., n=6, *P<0.05 for Gata4, BMP2 effectors and Hey1, 2 versus Gata4 and BMP2 effectors co-transfection, using two- and three-way ANOVA). (f) E9.5 chambers–Katushka/AV canal-EGFP transgenic hearts were cultured in the presence or absence of 50 μmol l−1 γ-secretase inhibitor (DAPT) for 48 h, and expression of AV canal and chamber markers was analysed with real time RT–PCR. Data were normalized to HPRT and expressed as folds of increase over untreated samples. (g) Untreated and DAPT-treated E9.5 hearts were subjected to ChIP assay analyses with anti-H3K27ac antibody. Pulled-down DNA fragments were analysed with RT–PCR and enrichment indicates the ratio of ChIPed DNA to negative control regions, normalized for input DNA. (h) Cartoon depicting the overlapping expressions of Hey1, 2 and Gata4 at stage E9.5–E10.5.