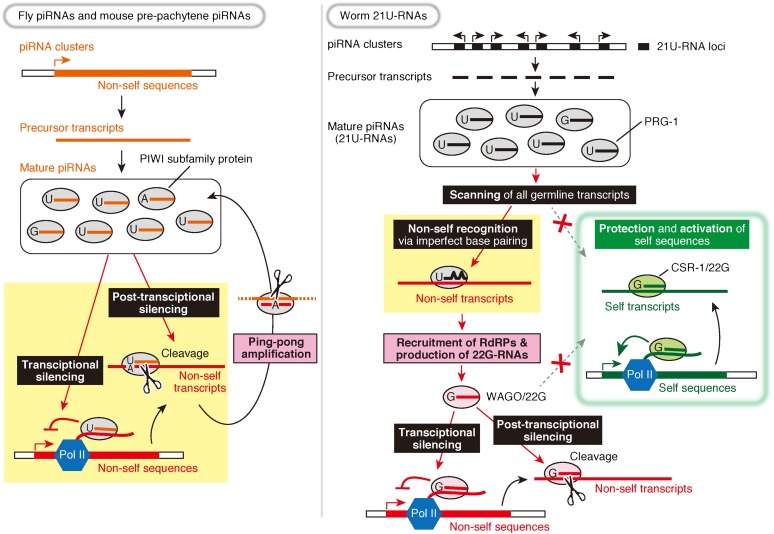
Figure 1.
Similarities and differences between the piRNA pathway in flies and mice and that in worms. Fly and mouse piRNA clusters generate long single-stranded piRNA precursor transcripts from which multiple piRNAs are produced. The mature piRNAs are loaded into PIWI proteins that target transposon RNAs with sequence complementarity and silence them by cleavage (post-transcriptional silencing). The cleaved transposon RNAs become new piRNA precursors and amplify piRNAs against the transposons (ping-pong amplification). A subset of PIWI proteins is localized to the nucleus and induces transcriptional silencing. In contrast, each 21U-RNA locus codes a single 21U-RNA precursor in worms. 21U-RNAs in PRG-1 scan virtually all germline transcripts to detect potential harmful exogenous sequences, allowing several mismatches for recognition. To avoid self-attack, the CSR-1/22G-RNA pathway marks endogenous protein-coding genes for their protection from silencing and their activation. When the PRG-1/21U-RNA complex recognizes its targets, RNA-dependent RNA polymerases (RdRPs) are recruited, and another class of 22G-RNAs is produced. These 22G-RNAs are loaded into worm-specific Ago proteins (WAGOs) and silence their targets at both transcriptional and post-transcriptional levels. The role of piRNAs is different between flies/mice and worms (highlighted in yellow), but both achieve the same purpose of nonself silencing. Note that worms lack the ping-pong amplification but instead have the WAGO/22G-RNA pathway, thereby amplifying silencing signals (highlighted in pink).