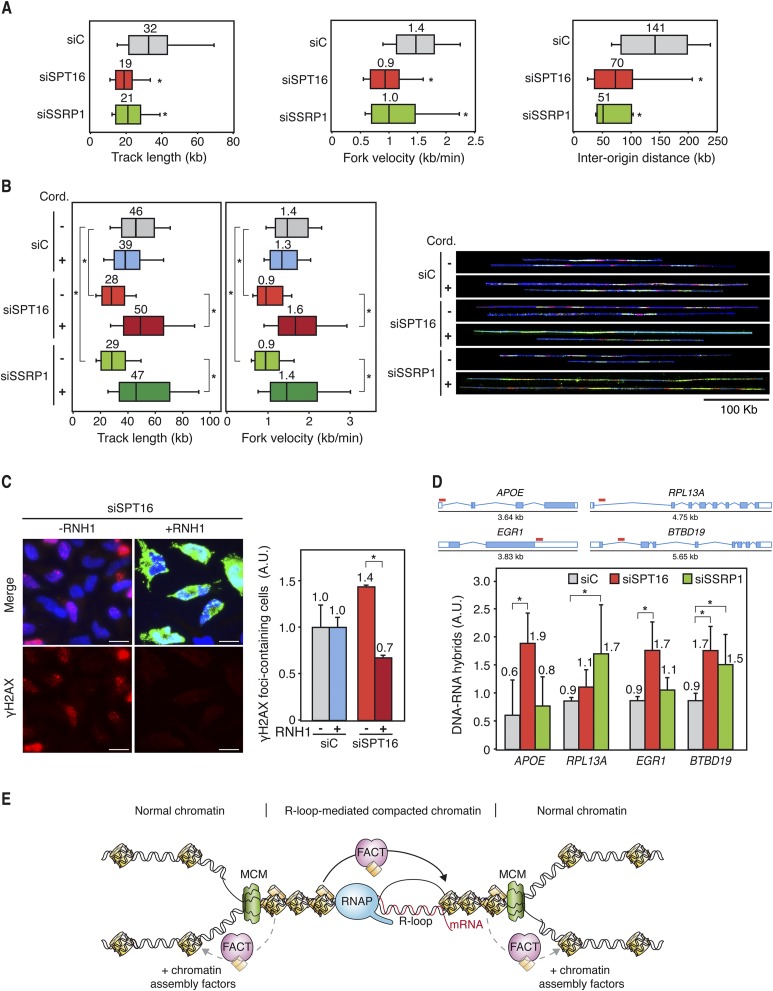
Figure 7.
Transcription-dependent replication deficiencies in hFACT-depleted cells. (A) Single-molecule analysis of DNA replication (DNA combing) in siC, siSPT16, and siSSRP1 transfected MRC-5 cells. Cells were pulse-labeled for 10 min with IdU followed by 20 min with CldU. Distributions of CldU track lengths, RF velocity, and interorigin distance are shown. Median values are indicated. Boxes and whiskers indicate 25–75 and 10–90 percentiles, respectively. (*) P < 0.05 (median test). (B) DNA combing in siC, siSPT16, and siSSRP1 transfected HeLa cells with or without 100 min of treatment with 50 μM cordycepin. Cells were pulse-labeled for 10 min with IdU (red) followed by 30 min with CldU (green). DNA is shown in blue. Other details are as in A. (C) Immunofluorescence of γH2AX in siC (control) and siSPT16 transfected HeLa cells with or without RNase H1 (RNH1) overexpression. Cells were transfected with pcDNA3 (control) or pcDNA3-RNaseH1 (RNH1) during 24 h. Nuclei were stained with DAPI. Bars, 25 μm. Graphics show the quantification of the percentage of cells containing γH2AX foci relative to the siC (control) in each case. Mean and SEM for two independent experiments are shown. (*) P < 0.05 (Student's t-test). (D) DRIP using the S9.6 antibody, which specifically recognizes DNA:RNA hybrids in siC, siSPT16, and siSSRP1 cells at APOE, RPL13A, EGR1, and BTBD19 genes. Signal values of R-loop detection normalized in respect to the siC control are shown. Signal values from samples treated with RNase H were subtracted. Red lines indicate the regions where DRIP analyses were performed. Mean and SD for four independent experiments are shown. (*) P < 0.05 (Mann-Whitney U-test). (E) Model to explain the molecular defects occurring in FACT-deficient cells. RF progression through R-loop-altered chromatin is facilitated by the FACT complex, which would assist nucleosome swapping to facilitate nucleosome reassembly at transcription–replication collisions sites. For clarity, only the MCM helicase is depicted at the RF.