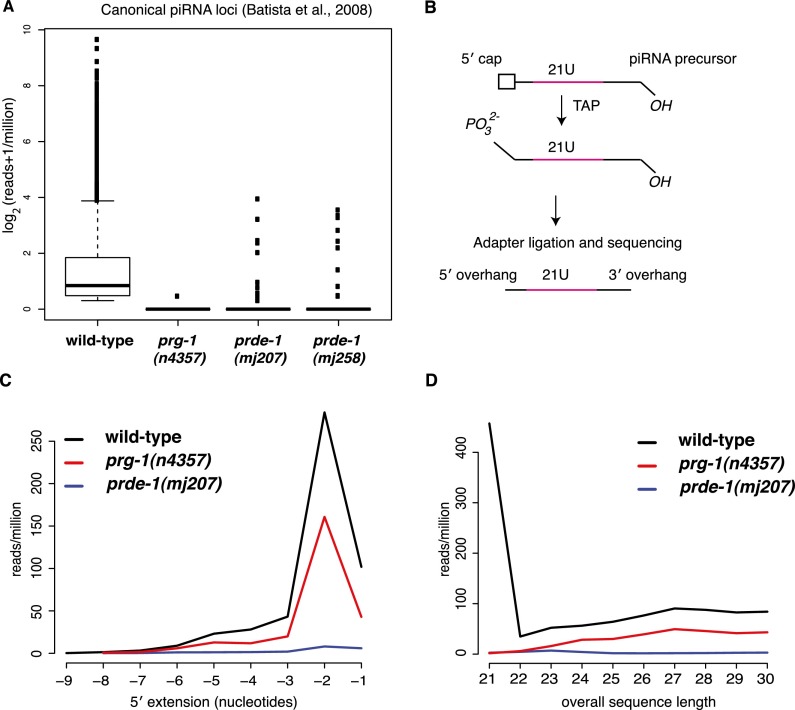
Figure 4.
PRDE-1 is required for piRNA precursor biogenesis. (A) 21U small RNA reads mapping perfectly to known C. elegans piRNA loci, as annotated previously (Batista et al. 2008), in prde-1 mutants compared with prg-1. 5′-dependent small RNA libraries were prepared from wild-type, prg-1(n4357), prde-1(mj207), and prde-1(mj258) RNA. Boxes represent interquartile range, with the median indicated by a line, and they extend to the maximum point no more than 1.5-fold greater than the interquartile range. Outliers are indicated by dots. (B) Outline of strategy for detecting potential piRNA precursors by deep sequencing. (C) Distribution of 5′ extending nucleotides for sequences mapping to piRNA sequences as used in A but that are >21 nt in wild type (black line), prg-1 (red line), and prde-1(mj207) (blue line). These are likely capped piRNA precursors. (D) Overall length of all sequences, which map perfectly to piRNA loci (i.e., either 21 nt in length or above) in wild type, prg-1, and prde-1. Color code is as in C.