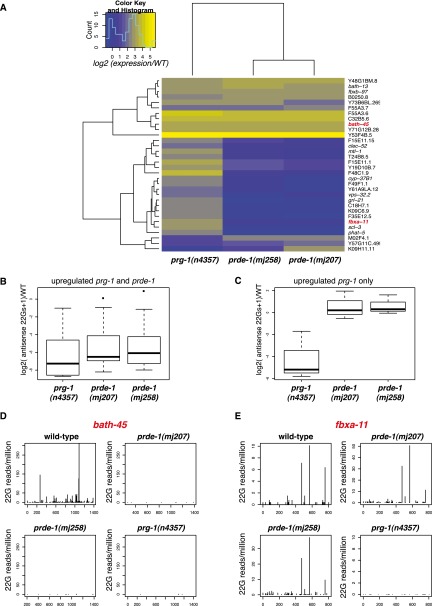
Figure 6.
PRDE-1-dependent and -independent piRNAs target a distinct set of genes. (A) Heat map showing the behavior of all genes that are statistically significantly up-regulated by more than fourfold with a P < 0.05, with a multiple test correction P < 0.1, in at least one out of prg-1 and the two prde-1 alleles relative to wild type. The color of each entry in the heat map shows the difference in mean expression relative to wild type (WT) as seen in the color key. (B,C) 22G-RNAs mapping to genes up-regulated in either prg-1 and prde-1 (B) or prg-1 alone (C). The number of antisense 22Gs relative to wild type, normalized to total library size, is shown on the Y-axis. Boxes represent interquartile range, with the median indicated by a line, and they extend to the maximum point no more than 1.5-fold greater than the interquartile range. Outliers are indicated by dots. (D,E) 22G-RNAs mapping to representative target genes of prde-1-dependent (D) and prde-1-independent (E) piRNAs. The start position of each 22G-RNA is plotted on the X-axis, with number of reads normalized to total library size on the Y-axis.