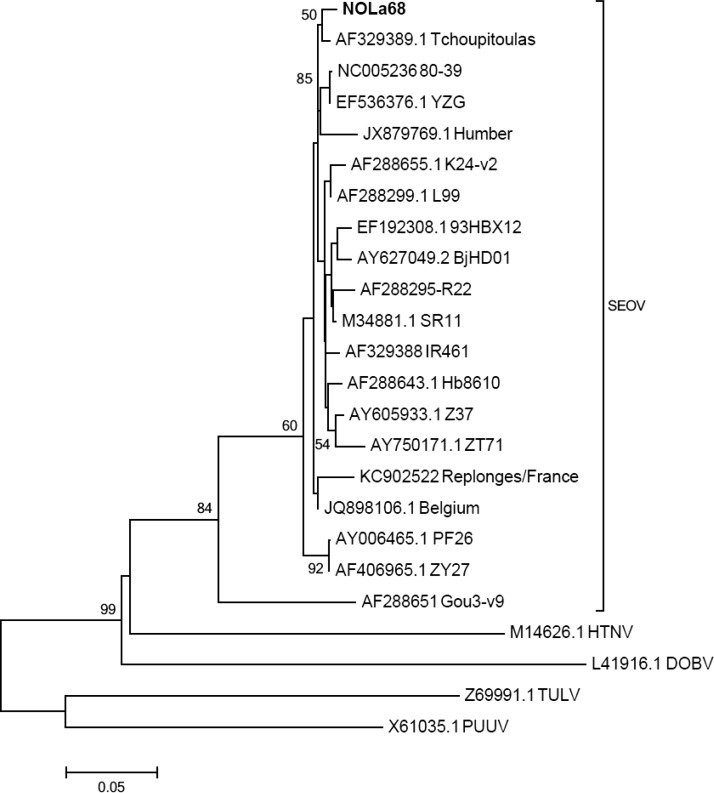
Figure 2.
Maximum likelihood phylogenetic tree showing relatedness of a representative hantavirus (NOLa68; bold) indentified from rodent samples collected in New Orleans, Louisiana, with other Old World hantavirus sequences available from GenBank. The tree compares partial S-segment sequences (nucleotide positions 973–1253 of accession number M14626.1) and was constructed using the Tamura three-parameter model with γ-distributed rate heterogeneity and a proportion of invariant sites (T92 + G + I). Each branch shows the GenBank accession number followed by common virus identifier. The scale bar indicates nucleotide substitutions per site. GenBank accession numbers of sequences derived from this study are KF683313, KF683314, and KF683315. DOBV = Dobrava virus; HTNV = Hantaan virus; PUUV = Puumala virus; SEOV = Seoul virus; TULV = Tula virus.