Table 1.
Proteins identified and analyzed by MALDI-TOF-TOF/MS
| Spot no | Protein name | Accession no. | Deduced apple genome ID | Protein MW/PI | Pep. count | Protein score | Relative protein content |
|---|---|---|---|---|---|---|---|
| Photosynthesis proteins | |||||||
| L8 |
ribulose-1,5-bisphosphate carboxylase |
gi|2961315 |
MDP0000597996 |
53314.4/6.14 |
5 |
253 |
 |
| L45 |
ribulose-1,5-bisphosphate carboxylase/oxygenase activase alpha 2 |
gi|78100212 |
MDP0000944409 |
46944.1/4.84 |
12 |
494 |
 |
| L49 |
ribulose-1,5-bisphosphate carboxylase/oxygenase activase small protein isoform |
gi|115334979 |
MDP0000321244 |
47952.1/7.57 |
10 |
326 |
 |
| L50 |
Oxygen-evolving enhancer protein 2, chloroplastic; Short = OEE2; |
gi|11134051 |
MDP0000361338 |
28674.3/6.6 |
4 |
231 |
 |
| L51 |
ribulose 1,5-bisphosphate carboxylase |
gi|4206530 |
MDP0000597996 |
49888.8/5.94 |
3 |
223 |
 |
| L82 |
ribulose 1,5-bisphosphate carboxylase |
gi|10945633 |
MDP0000597996 |
52133/6.18 |
8 |
154 |
 |
| L85 |
ribulose-phosphate 4-epimerase |
gi|225457361 |
MDP0000137234 |
30146/8.93 |
5 |
533 |
 |
| L95 |
ribulose 1,5-bisphosphate carboxylase |
gi|371928199 |
MDP0000597996 |
49673/6.04 |
20 |
498 |
 |
| L15 |
Chlorophyll a-b binding protein 3C-like protein |
gi|357497757 |
MDP0000784451 |
24884.4/5.53 |
4 |
133 |
 |
| L19 |
putative chlorophyll a/b binding protein |
gi|397789264 |
MDP0000875642 |
16135/7.82 |
6 |
262 |
 |
| L61 |
Chlorophyll a-b binding protein 2, chloroplastic; |
gi|158562858 |
MDP0000708928 |
3798.9/8.2 |
4 |
212 |
 |
| L87 |
chlorophyll a-b binding protein 8, chloroplastic |
gi|225436257 |
MDP0000866655 |
29511.2/7.85 |
2 |
192 |
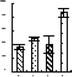 |
| Amino acid metabolism related proteins | |||||||
| L43 |
Serine-type peptidase |
gi|357495999 |
MDP000065459 |
45945.4/6.79 |
9 |
267 |
 |
| L54 |
precursor of carboxylase p-protein 1, glycine decarboxylase complex |
gi|224088838 |
MDP0000588069 |
115985/6.51 |
11 |
407 |
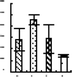 |
| L58 |
putative plastidic glutamine synthetase |
gi|26892040 |
MDP0000139493 |
47619.1/6.77 |
9 |
457 |
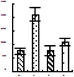 |
| Transpot related proteins | |||||||
| L56 |
Transketolase, chloroplastic |
gi|75140229 |
MDP0000142098 |
73346.7/5.47 |
6 |
20-6 |
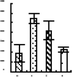 |
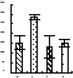
| L81 |
alanine aminotransferase 3 |
gi|351724777 |
MDP0000815368 |
53789.3/5.52 |
11 |
470 |
 |
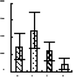
| L99 |
PREDICTED: transketolase, chloroplastic-like |
gi|356536526 |
MDP0000142098 |
80689.8/6.12 |
11 |
299 |
 |
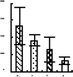
| L101 |
Transketolase |
gi|357445031 |
MDP0000142098 |
80087.5/6.00 |
10 |
219 |
 |
| Energy metabolism related proteins | |||||||
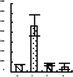
| L23 |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic-like isoform 1 |
gi|356538289 |
MDP0000811918 |
40803.8/8.7 |
14 |
477 |
 |
| L24 |
NAD dependent epimerase/dehydratase |
gi|255559448 |
MDP000019372 |
43230.8/8.75 |
6 |
243 |
 |
| Carbohydrate metabolism related proteins | |||||||
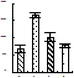
| L27 |
PREDICTED: triosephosphate isomerase |
gi|225449541 |
MDP0000152242 |
27396.2/6.34 |
5 |
373 |
 |
| L33 |
alcohol dehydrogenase |
gi|307135978 |
MDP0000239956 |
40657.6/8.53 |
3 |
138 |
 |
| L36 |
cytosolic malate dehydrogenase |
gi|78216493 |
MDP0000174740 |
35970.5/6.01 |
3 |
352 |
 |
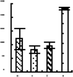
| L40 |
glyceraldehyde-3-phosphate dehydrogenase A |
gi|381393060 |
MDP0000527995 |
43224.5/8.1 |
18 |
853 |
 |
| L41 |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic |
gi|225460680 |
MDP0000251810 |
37554/6.03 |
7 |
332 |
 |
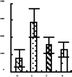
| L42 |
glyceraldehyde-3-phosphate dehydrogenase B |
gi|381393062 |
MDP0000835914 |
48490.9/8 |
21 |
790 |
 |
| L46 |
glyceraldehyde 3-phosphate dehydrogenase A subunit |
gi|62318887 |
MDP0000527995 c |
17730.1/5.35 |
6 |
210 |
 |
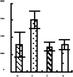
| L47 |
NADP-dependent glyceraldehyde-3-phosphate dehydrogenase; |
gi|3913711 |
MDP0000152497 |
53654.6/6.76 |
10 |
375 |
 |
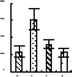
| L53 |
alcohol dehydrogenase |
gi|307135978 |
MDP0000239956 |
40657.6/8.53 |
8 |
216 |
 |
| L59 |
chloroplast sedoheptulose-1,7-bisphosphatase |
gi|118175929 |
MDP0000244771 |
42824.9/6.06 |
9 |
377 |
 |
| L83 |
PREDICTED: triosephosphate isomerase |
gi|225449541 |
MDP0000152242 |
27396.2/6.34 |
6 |
266 |
 |
| L86 |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta |
gi|357159289 |
MDP0000146411 |
40081.6/5.47 |
5 |
407 |
 |
| Binding proteins | |||||||
| L68 |
ATP synthase, subunit B |
gi|7578491 |
MDP0000565338 |
53365/5.57 |
15 |
752 |
 |
| L69 |
ATP synthase subunit B |
gi|91983091 |
MDP0000565338 |
51812.1/5.27 |
21 |
703 |
 |
| L71 |
ATP synthase, subunit B |
gi|7578491 |
MDP0000565338 |
53365/5.57 |
19 |
791 |
 |
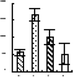
| L72 |
putative ATP synthase subunit B |
gi|56784991 |
MDP0000565338 |
45936.9/5.33 |
17 |
971 |
 |
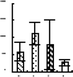
| L73 |
PREDICTED: ATP synthase subunit B, mitochondrial-like |
gi|225424142 |
MDP0000565338 |
59699.2/5.84 |
18 |
760 |
 |
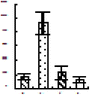
| L77 |
ATP synthase, subunit B |
gi|7578491 |
MDP0000565338 |
53365/5.57 |
19 |
632 |
 |
| L78 |
ATP synthase CF1 alpha subunit |
gi|346683279 |
MDP000092905 |
55443.9/5.09 |
21 |
1100 |
 |
| L79 |
ATP synthase subunit B |
gi|91983091 |
MDP0000565338 |
51812.1/5.27 |
24 |
923 |
 |
| Antioxidant related proteins | |||||||
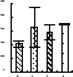
| L25 |
Probable aldo-keto reductase 2 |
gi|378548276 |
MDP0000228499 |
38436.6/6.17 |
6 |
341 |
 |
| L37 |
aldo/keto reductase AKR |
gi|62526573 |
MDP0000228499 |
38026.5/6.38 |
10 |
483 |
 |
| Defense and stress related proteins | |||||||
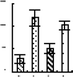
| L13 |
class III endo-chitinase |
gi|33347391 |
MDP0000280265 |
17774.6/4.12 |
1 |
207 |
 |
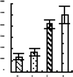
| L16 |
heat shock protein 70 |
gi|30962610 |
MDP0000322220 |
47584/5.1 |
9 |
106 |
 |
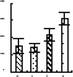
| L17 |
beta-1,3-glucanase |
gi|399137110 |
MDP0000570395 |
37551.2/5.07 |
4 |
258 |
 |
| L18 |
thioredoxin peroxidase |
gi|21912927 |
MDP000020081 |
30103.4/8.2 |
5 |
328 |
 |
| L22 |
thaumatine-like protein |
gi|20149274 |
MDP0000552328 |
22948.5/4.6 |
7 |
317 |
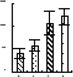 |
| L28 |
ascorbate peroxidase |
gi|145581388 |
MDP0000199034 |
27711.8/5.53 |
6 |
414 |
 |
| L29 |
ascorbate peroxidase |
gi|319993039 |
MDP0000194474 |
12064.3/8.72 |
3 |
73 |
 |
| L30 |
early-responsive to dehydration 2 |
gi|383100964 |
MDP0000322220 |
69263.1/5.19 |
11 |
149 |
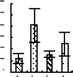 |
| L84 |
quinone oxidoreductase [Gymnadenia conopsea] |
gi|89276317 |
MDP0000393227 |
21631.1/6.06 |
2 |
159 |
 |
| L89 |
phi class glutathione S-transferase protein |
gi|329130898 |
MDP0000266097 |
23996.3/5.97 |
3 |
90 |
 |
| L90 |
IgE-binding protein MnSOD |
gi|10862818 |
MDP0000187714 |
22957.7/6.06 |
6 |
397 |
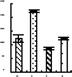 |
| Unknown proteins | |||||||
| L20 |
uncharacterized protein LOC100805310 |
gi|359806638 |
MDP0000875642 |
27959.1/5.29 |
9 |
353 |
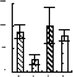 |
| L31 |
PREDICTED: uncharacterized protein At2g37660, chloroplastic |
gi|225440390 |
MDP0000176370 |
27681.5/5.85 |
2 |
150 |
 |
| L32 |
predicted protein |
gi|224087915 |
MDP0000641719 |
27403.9/5.74 |
9 |
464 |
 |
| L38 |
PREDICTED: clavaminate synthase-like protein At3g21360 |
gi|225442460 |
MDP0000406399 |
36588.7/6.1 |
4 |
80 |
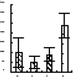 |
| L57 | predicted protein | gi|224138316 | MDP0000148186 | 45396.1/6.11 | 14 | 534 |  |
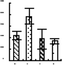
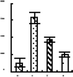
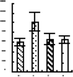
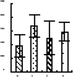
*In relative protein content graph, from left to right were  : Control group 3 days post-treatment,
: Control group 3 days post-treatment,  : Inoculated group 3 days post-treatment,
: Inoculated group 3 days post-treatment,  : Control group 6 days post-treatment,
: Control group 6 days post-treatment,  : Inoculated group, 6 days post-treatment respectively.
: Inoculated group, 6 days post-treatment respectively.
