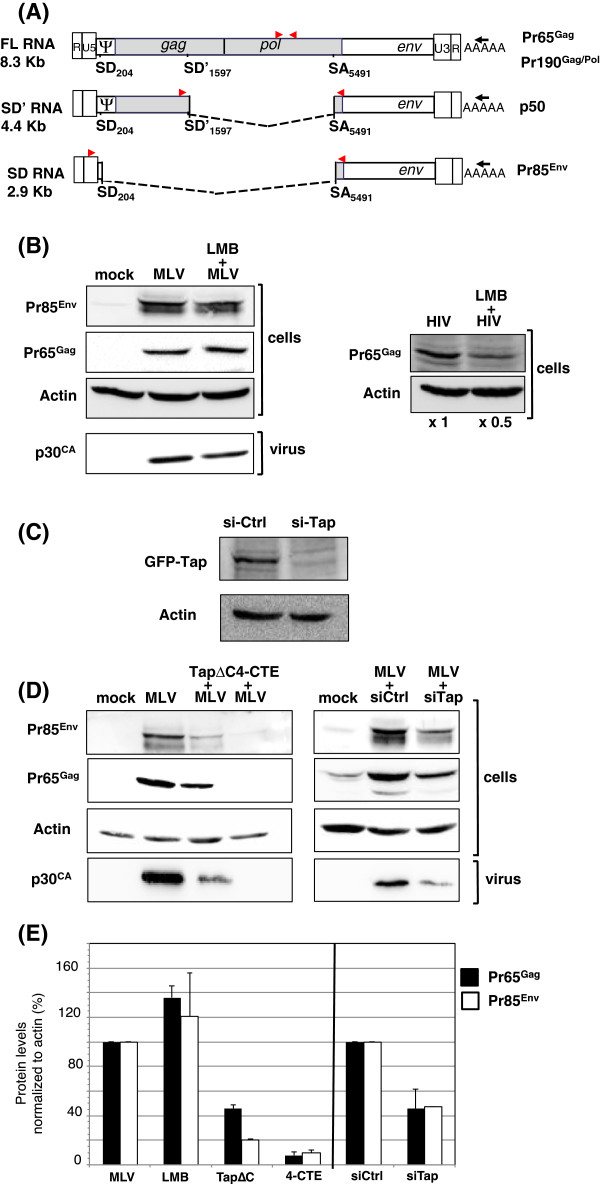
Figure 1.
Effects of CRM1 and Tap inhibition on MLV expression. (A) RNAs produced by MLV and their encoded proteins. Black arrows correspond to the RT primers while positions of the RT-qPCR primers used to amplify the different RNAs are indicated by red arrows. (B) Investigation of the CRM1 pathway by Western blot analysis of MLV or HIV proteins produced in LMB-treated NIH3T3 or 293 T cells, respectively. Cells were transfected with 2 μg of pMov9.1 or pNL4.3 plasmid, the molecular clones of Mo-MLV and HIV-1 respectively, for 48 h and incubated for the last 8 h with 20 nM LMB (LC Laboratories) before harvesting. (C) Activity of the Tap siRNA. 1 μg of TAPΔC was cotransfected in NIH3T3 cells with 100 pmol of Tap Stealth RNAi (invitrogen) or control siRNA (Eurogentec). Cell were collected 48 h p.t. and Tap expression analyzed by Western blotting with anti-GFP antibodies (Roche). (D) Investigation of the Tap pathway by Western blot analysis of MLV proteins produced in NIH3T3 cells transfected for 48 h with 1 μg pMov9.1 with either 1 μg of peGFP-TapΔC [19], 1.5 μg of p3-CCCC (4-CTE) [18] or 1.5 μg of empty pcDNA3.1 vector. In all experiments, MLV:inhibitor ratios are 1:3 for peGFP-TapΔC and p3-CCCC (or pcDNA3.1). For RNA interference, NIH3T3 cells were co-transfected for 48 h with 2 μg pMov9.1 and 100 pmol of either Tap stealth RNAi as described in [18], or control siRNA. Viruses were collected by ultracentrifugation from culture supernatant and loaded on SDS-PAGE. Procedure and antibodies used to detect viral proteins are described in [16]. Actin was detected with a rabbit polyclonal antibody (1/300) (Sigma) and a peroxidase-conjugated (HRP) goat anti-rabbit antibody (1:4000) (Sigma). (E) Band integrated densities were determined with the ImageJ software and normalized to actin. Intensities are presented as mean +/− SEM.