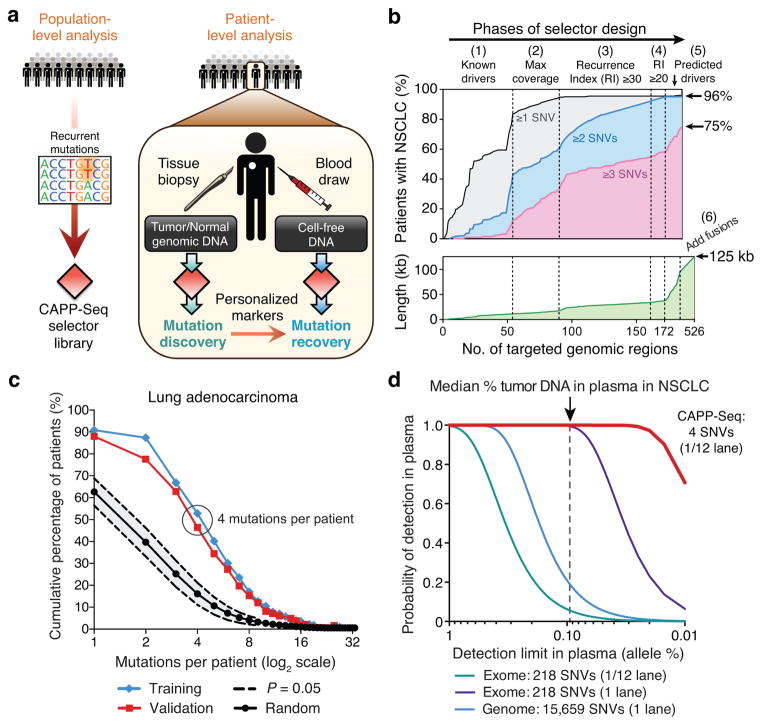
Figure 1. Development of CAncer Personalized Profiling by deep Sequencing (CAPP-Seq).
(a) Schematic depicting design of CAPP-Seq selectors and their application for assessing circulating tumor DNA. (b) Multi-phase design of the NSCLC selector. Phase 1: Genomic regions harboring known and suspected driver mutations in NSCLC are captured. Phases 2–4: Addition of exons containing recurrent SNVs using WES data from lung adenocarcinomas and squamous cell carcinomas from TCGA (n = 407). Regions were selected iteratively to maximize the number of mutations per tumor while minimizing selector size. Recurrence index = total unique patients with mutations covered per kb of exon. Phases 5,6: Exons of predicted NSCLC drivers15,16 and introns and exons harboring breakpoints in rearrangements involving ALK, ROS1, and RET were added. Bottom: increase of selector length during each design phase. (c) Analysis of the number of SNVs per lung adenocarcinoma covered by the NSCLC selector in the TCGA WES cohort (Training; n = 229) and an independent lung adenocarcinoma WES data set (Validation; n = 183)20. Results are compared to selectors randomly sampled from the exome (P < 1.0 × 10−6 for the difference between random selectors and the NSCLC selector). (d) Analytical modeling of CAPP-Seq, whole exome and whole genome sequencing for different detection limits of tumor circulating DNA in plasma. Calculations are based on the median number of mutations detected per NSCLC for CAPP-Seq (i.e., 4) and the reported number of mutations in NSCLC exomes and genomes21. Additional details are described in Methods. The vertical dotted line represents the median fraction of tumor-derived circulating DNA detected in plasma from patients in this study.