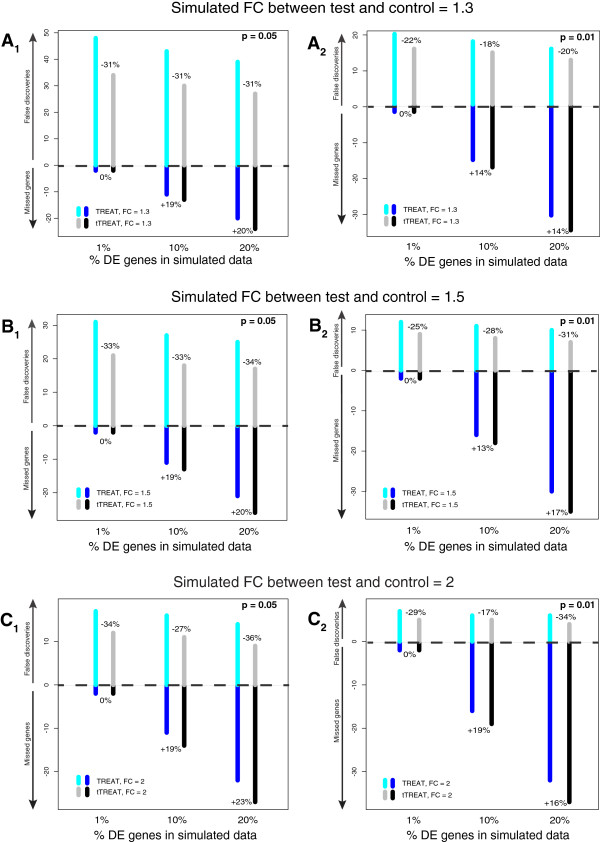
Figure 1.
False discoveries and missed genes for TREAT and tTREAT on simulated data, at p = 0.05 or at p = 0.01. (A1) The positive y axis shows the average of the false discoveries, and the negative y axis shows the average of the missed genes for TREAT and tTREAT on 400 simulated datasets. The x axis shows the percentage of DE genes (1%, 10% and 20%) that is simulated in each case. The data are simulated with respect to a FC difference ω of 1.3 (up or down), and the FC threshold τ used for TREAT and tTREAT is also 1.3. The percentages next to the gray and black bars of tTREAT represent the percentual decrease (prefixed with a minus sign) or increase (prefixed with a plus sign) in false discoveries or missed genes with respect to the reference, TREAT (depicted in cyan and blue). The significance cut-off was set at p = 0.05. (A2) Same conditions as in panel A1, except that significance was set at p = 0.01. (B1) Similar to panel A1, but now the data are simulated with respect to a FC difference ω of 1.5, and the FC threshold τ used for TREAT and tTREAT is also 1.5. The significance cut-off was set at p = 0.05. (B2) Same conditions as in panel B1, except that significance was set at p = 0.01. (C1) Similar to panel A1, but now the data are simulated with respect to a FC difference ω of 2, and the FC threshold τ used for TREAT and tTREAT is also 2. The significance cut-off was set at p = 0.05. (C2) Same conditions as in panel C1, except that significance was set at p = 0.01.