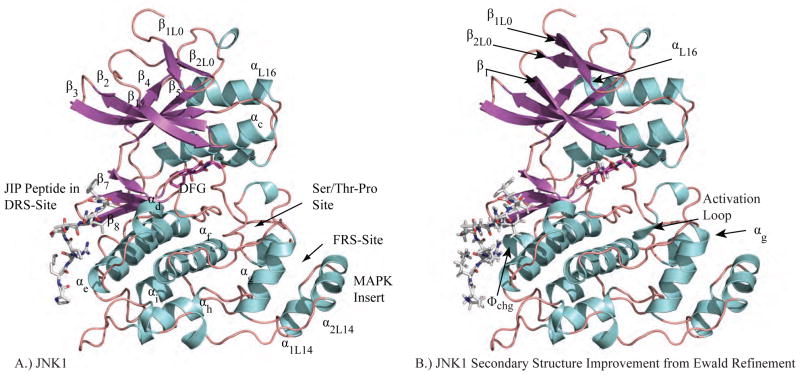
Figure 2.
A.) JNK1 (PDB ID: 3O17) is shown with each segment of secondary structure labeled. The subscripts for secondary structure elements that are not conserved for all protein kinases refer to loop number. For example, the first β-strand of Loop 0 is β1L0. B.) Ewald refinement of the original experimental data using prior chemical information defined by the polarizable AMOEBA force field improves the quality of the structural model via extension of energetically favorable secondary structure elements and by improving agreement with the measured diffraction data (ie. reducing Rfree from 30.6 to 27.9). The 3 β-strands and 4 α-helices that were extended are labeled. Changes near the DRS-site (Φchg) and FRS-site (αg, activation loop) may impact downstream computational methods such as virtual screening of non-ATP MAP kinase inhibitors.