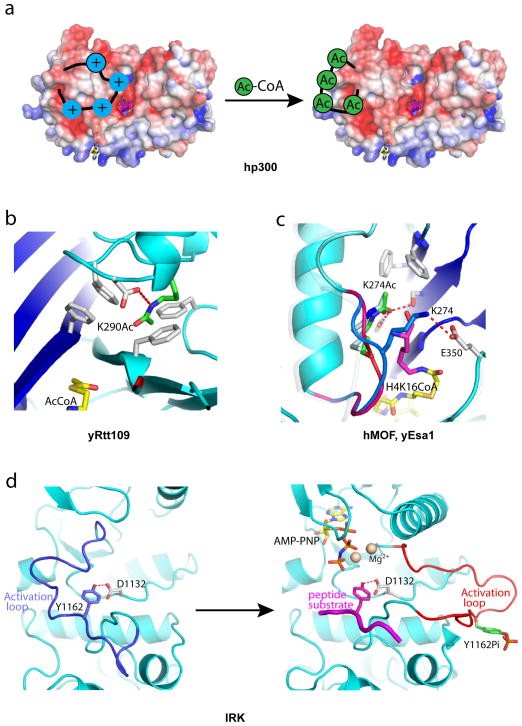
Figure 4. Auto Regulation of HAT Proteins and Kinases.
Close-up views of the autoacetylation site of HAT proteins and kinases. (a) Model for hp300 activation by autoacetylation (electrostatic potential maps are contoured at ± 4.0 kT/e, the same as in Figure 3), (b) The Lys290 autoacetylation site of yRtt109 highlighting the environment around acetylated Lys290. (c) Structure of hMOF highlighting the environment around acetylated Lys274 (green stick). The loop harboring Lys274 is highlighted in red for the acetylated conformation and blue for the unacetylated conformation. Superposition of the yEsa1/H4K16CoA structure with hMOF indicates that the unacetylated conformation would clash with binding of the cognate substrate lysine (as represented by the lysine of the H4K16CoA bisubstrate inhibitor shown in purple). (d) Structures of insulin receptor tyrosine kinase showing the autoinhibited conformation (left, PDB 1IRK) and autoactivated conformation (right, PDB 1IR3). The activation loop harboring unphosphorylated Tyr1162 adopts an autoinhibited conformation (blue) that would clash with the binding of both the cofactor (yellow stick) and cognate peptide substrate (purple).