Abstract
In medical imaging analysis and computer vision, there is a growing interest in analyzing various manifold-valued data including 3D rotations, planar shapes, oriented or directed directions, the Grassmann manifold, deformation field, symmetric positive definite (SPD) matrices and medial shape representations (m-rep) of subcortical structures. Particularly, the scientific interests of most population studies focus on establishing the associations between a set of covariates (e.g., diagnostic status, age, and gender) and manifold-valued data for characterizing brain structure and shape differences, thus requiring a regression modeling framework for manifold-valued data. The aim of this paper is to develop an intrinsic regression model for the analysis of manifold-valued data as responses in a Riemannian manifold and their association with a set of covariates, such as age and gender, in Euclidean space. Because manifold-valued data do not form a vector space, directly applying classical multivariate regression may be inadequate in establishing the relationship between manifold-valued data and covariates of interest, such as age and gender, in real applications. Our intrinsic regression model, which is a semiparametric model, uses a link function to map from the Euclidean space of covariates to the Riemannian manifold of manifold data. We develop an estimation procedure to calculate an intrinsic least square estimator and establish its limiting distribution. We develop score statistics to test linear hypotheses on unknown parameters. We apply our methods to the detection of the difference in the morphological changes of the left and right hippocampi between schizophrenia patients and healthy controls using medial shape description.
1 Introduction
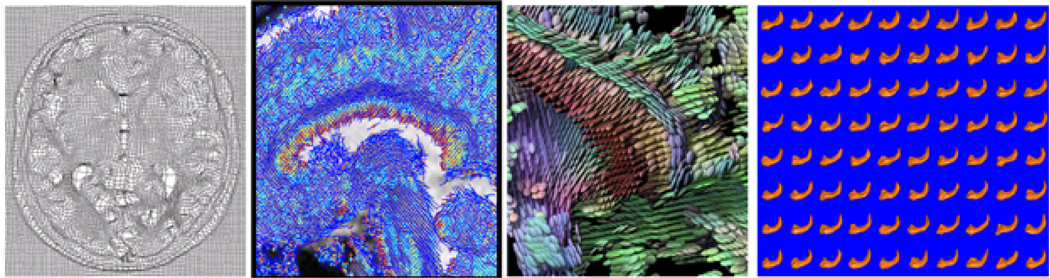
Statistical analysis of manifold-valued data has gained a great deal of attention in neuroimaging applications [1], [2], [3], [4], [5], [6], [7], [8], [9]. Examples of manifold-valued data that we encounter in medical imaging analysis include the Grassmann manifold, planar shapes, deformation field, symmetric positive definite (SPD) matrices and the medial shape representations (m-rep) of subcortical structures (Fig. 1). Some review papers on the applications of manifold-valued data in medical imaging can be found in a recent special issue of NeuroImage [10]. However, the existing statistical methods for manifold-valued data are primarily developed to estimate intrinsic and extrinsic means, to estimate the structure of population variability, to carry out principal geodesic analysis, and to compare intrinsic (or extrinsic) means across two or more groups [11], [9], [12], [13], [14], [15], [16], [17].
Fig. 1.
Four different manifold-valued data (from the left to the right): deformation field reflecting brain deformation obtained from the registration of either diffusion tensor images (DTIs) or T1 magnetic resonance images (T1 MRIs); principal direction (PD) field reflecting fiber orientations obtained from DTIs; diffusion tensor field reflecting water diffusion along fiber tracts from DTIs; medial shape representations of hippocampi from multiple subjects obtained from the segmented T1 MRIs
Little literature exists for regression analyses of manifold-valued data. The existing parametric and nonparametric regression models for manifold-valued data were primarily developed for 2 (or 3) dimensional directional data [6], [18]. In parametric regression of directional data, parametric distributions, such as the Von Mises distribution, are commonly assumed for directional data, whereas it can be very challenging to assume useful parametric distributions for other manifold-valued data, such as SPD matrices and the m-rep, which can characterize the feature (e.g., shape) of real imaging data [15]. In the nonparametric analysis of manifold-valued data, although smoothing splines have been developed for directional data and planar landmark data, it is computationally difficult to generalize such smoothing splines to other manifold-valued data [6]. Recently, local constant regressions have been developed for manifold-valued data, but these regression models are defined with respect to either the Frechet mean or the geometric median [2], [4].
According to the best of our knowledge, this is the first paper that develops a semiparametric regression model with manifold-valued data as responses on a Riemannian manifold and a set of covariates, such as time, gender, and diagnostic status, in Euclidean space. Our regression model are solely based on the first-order moment, thus avoiding specifying any parametric distributions. We propose an inference procedure to estimate the regression coefficients in this semi-parametric model. We establish asymptotic properties, including consistency and asymptotic normality, of the estimates of the regression coefficients. We develop score statistics to test linear hypotheses on unknown parameters. Finally, we illustrate the application of our statistical methods to the detection of the difference in morphological changes of the hippocampi between schizophrenia patients and healthy controls in a neuroimaging study of schizophrenia.
2 Method
2.1 Review of Regression Models
We consider a dataset that is composed of a response yi and a p × 1 covariate vector xi for i = 1, ⋯, n. Responses may be continuous observations in classical linear models, such as age, weight, income, and they may be discrete or ordinal observations, such as differing severity of diseases and disease status (patients v.s. healthy subjects). Covariates may be quantitative, such as age, or qualitative, such as handiness, gender, and presence of risk factors (yes/no).
Regression models often include two key elements: a link function μi(β) = g(xi, β) and a residual εi = yi − μi(β), where β is a q × 1 vector of regression coefficients and g(·,·) is an known mapping from Rp × Rq to R. Regression models assume that
| (1) |
where the expectation denotes the conditional expectation of ε given x. Moreover, nonparametric regressions include a link function μi = g(xi), in which g(·) is an unknown function, and a residual εi = yi − g(xi), for which equation (1) holds.
To carry out statistical inference on β (or g(·)), we need at least three statistical methods. The first one is an estimation method for calculating parameter estimate of β, denoted by β̂. Various estimation methods include maximum likelihood estimation, robust estimation, estimating equations, among many others. The second is to prove that β̂ is a consistent estimator of β and has certain asymptotic distribution (e.g., normal). The third is to develop test statistics for testing the hypotheses:
| (2) |
where h0(·) is an r × 1 vector function and b0 is an r × 1 specified vector. In most applications, we are interested in testing h0(β) = Hβ = b0 for a given r × q matrix H [18], [7], [8].
2.2 Intrinsic Regression for Manifold-Valued Data
We formally develop an intrinsic regression model for manifold-valued responses and covariates of interest from n observations. Suppose we observe a dataset {(Si, xi) : i = 1, ⋯, n}, where Si are points on a Riemannian manifold 𝓢 and xi are covariates of interest in Euclidean space.
The intrinsic regression model first involves modeling a ‘conditional mean’ of an manifold-valued response Si given xi, denoted by μi(β) = g(xi, β), where g(·,·), called link function, is a map from Rp × Rq to the manifold 𝓢. Note that we just borrow the term ‘conditional mean’ from Euclidean space. Given two points Si and μi(β) on the manifold ℳ, the intrinsic regression model also define the residual or ‘difference’ between Si and μi(β) to ensure that μi(β) is the proper ‘conditional mean’ of Si given xi. At μi(β), we have a tangent space of the manifold 𝓢, denoted by Tμi(β)𝓢, which is a Euclidean space representing a first order approximation of the manifold 𝓢 near μi(β). Then, we calculate the projection of Si onto Tμi(β)𝓢, denoted by Logμi(β)(Si), which can be regarded as the difference between Si and μi(β) for i = 1, ⋯, n. If 𝓢 is a Euclidean space, then Logμi(β)(Si) = Si − μi(β).
The intrinsic regression model for manifold-valued data is then defined by
| (3) |
for i = 1, ⋯, n, where the expectation is taken with respect to the conditional distribution of Si given xi. Model (3) does not assume any parametric distribution for Si given xi, and thus it allows for a large class of distributions [15]. In addition, our model (3) does not assume homogeneous variance across all i. This is also desirable for the analysis of imaging measures, such as diffusion tensors, because between-subject and between-voxel variability in the imaging measures can be substantial.
2.3 Estimation
We calculate an intrinsic least squares estimator (ILSE) of the parameter vector β, denoted by β̂, by minimizing the total residual sum of squares given by
| (4) |
where << ·,·>> is an inner product of two tangent vectors in Tμi(β)𝓢 and d(·,·) is the Riemannian distance function on 𝓢. Thus, let , β̂ solves the estimating equations given by
| (5) |
where ∂ denotes partial differentiation with respect to a parameter vector, such as β. The ILSE is closely related to the intrinsic mean μ̂IM of S1, ⋯, Sn ∈ 𝓢, which is defined as
| (6) |
In this case, μi is independent of i and covariates of interest. Moreover, under some conditions, we can establish consistency and asymptotically normality of β̂. A Newton-Raphson algorithm is developed to obtain β̂. Let ∂βGn(β) and , respectively, be the first- and second-order partial derivatives of Gn(β). We iterates , where 0 < ρ = 1/2k0 ≤ 1 for some k0 ≥ 0 is chosen such that Gn(β(t+1)) ≤ Gn(β(t)). We stop the Newton-Raphson algorithm when the absolute difference between consecutive β(t)’s is smaller than a predefined small number, say 10−4. Finally, we set β̂ = β(t). In addition, because may not be positive definite, we use instead of in order to stabilize the Newton-Raphson algorithm.
2.4 Hypotheses and Test Statistics
In medical analysis, most scientific questions of interest involve a comparison of manifold-valued data across diagnostic groups or detecting change in manifold-valued data across time [8], [19]. These scientific questions usually can be formulated as follows:
| (7) |
We test the null hypothesis H0 : Hβ = b0 using a score test statistic Wn defined by
| (8) |
where and , in which β̃ denotes the estimate of β under H0 and Ûi(β̃) is associated with ∂βGn(β). It can be shown that Wn is asymptotically χ2 distributed.
2.5 Positive Definitive Matrices
We develop an intrinsic regression for SPDs. We introduce the tangent vector and tangent space at any μ ∈ Sym+ (m), the space of SPDs [8]. The tangent space of Sym+ (m) at μ, denoted by TμSym+ (m), is identified with a copy of Sym(m), the space of symmetric matrices. Then we consider the scaled Frobenius inner product of any two tangent vectors Yμ and Zμ in TμSym+(m), which is defined by << Yμ, Zμ >>= tr(Yμμ−1Zμμ−1). Given the inner product, we can formally construct the Riemannian geometry of Sym+(m) [8].
We consider the link function μ(x, β) using the Cholesky decomposition of μ(x, β). For the i−th observation, through a lower triangular matrix Ci(β) = C(xi, β) = (Cjk(xi, β)), the Cholesky decomposition of μ(xi, β) equals μ(xi, β) = μi(β) = Ci(β)Ci(β)T. We must specify the explicit forms of Cjk(xi, β) for all j ≥ k in order to determine all entries in μi(β). As an illustration, for m = 2, we may choose the 2 × 2 matrix Ci(β) with , C12(xi, β) = 0, , and , where and β(k) for k = 1, 2,3 are subvectors of β. We introduce a definition of ‘residuals’ to ensure that μi(β) is the proper ‘conditionalmean’ of Si given xi. Then, we calculate the residual Logμi(β)(Si) given by Ci(β) log(Ci(β)−1SiCi(β)−T)Ci(β)T. The intrinsic regression is defined in (3).
The first- and second-order derivatives of Gn(β) are given as follows. The a-th element of ∂βGn(β) is given by , where ℇi(β) = log(Ci(β)−1SiCi(β)−T) and ∂βa = ∂/∂βa. The (a, b)-th element of is given by where and , in which
| (9) |
and S̃i(β) = Ci(β)−1SiCi(β)−T.
2.6 Median Representation
We develop an intrinsic regression for m-reps. An m-rep model consisting of k medial atoms can be considered as the direct product of k copies of M(1) = R3 × R+ × S(2) × S(2), that is , where S(2) is the sphere in R3 with radius one [5]. We introduce a tangent space TPM(1) at the point P = (O, r, n0, n1), where O ∈ R3, r ∈ R+, and n0 and n1 ∈ S(2). The tangent vector U ∈ TPM(1) takes the form U = (U0, Ur, Un0, Un1), where U0 ∈ R3, Ur ∈ R, Uni ∈ R3 and for i = 0, 1. The inner product of any two tangent vectors U(0) and U(1) in TPM(1) is defined by << U(0), U(1) >>= U(0)TU(1). The geodesic distance between P and P1 = (O1, r1, n0,1, n1,1) in M(1) is uniquely given by
To introduce an intrinsic regression for m-rep, we need to define a link function μ(x, β) = (μO(x, β), μr(x, β), μ0(x, β), μ1(x, β))T ∈ M(1), which is a 10 × 1 vector. For instance, we may set μO(x, β) = (xTβ1, xT β2, xTβ3)T and μr(x, β) = exp(xTβ4). A link function for is based on the stereographic projection given by
| (10) |
where g5(·) and g6(·) are known link functions and β5,k and β6,k are subvectors of β. The residual Logμ(x, β)(P) is given by
where Logμ0(x, β)(Un0) = arccos(μ0(x, β)TUn0)v/||v||2, in which v = Un0 − (μ0(x, β)TUn0)μ0(x, β).
3 Results
To demonstrate our regression method, we applied our methods to the m-rep shape of the hippocampus structure in the left and right brain hemisphere in schizophrenia patients and healthy controls, collected at 14 academic medical centers in North America and western Europe [19]. There were 56 healthy controls and 238 schizophrenia patients who met the following criteria: age 16 to 40 years; onset of psychiatric symptoms before age 35; diagnosis of schizophrenia, schizophreniform, or schizoaffective disorder according to DSM-IV criteria; and various treatment and substance dependence conditions.
We investigated the difference of m-rep shape between schizophrenia patients and healthy controls while controlling for other factors, such as gender and age. The hippocampi m-rep shape at the 24 medial atoms of the left and right brain were used as the response in our intrinsic regression model. Covariates of interest include Whole Brain Volume (WBV), race (Caucasian, African American and others), age (in years), gender, and diagnostic status (patient or control).
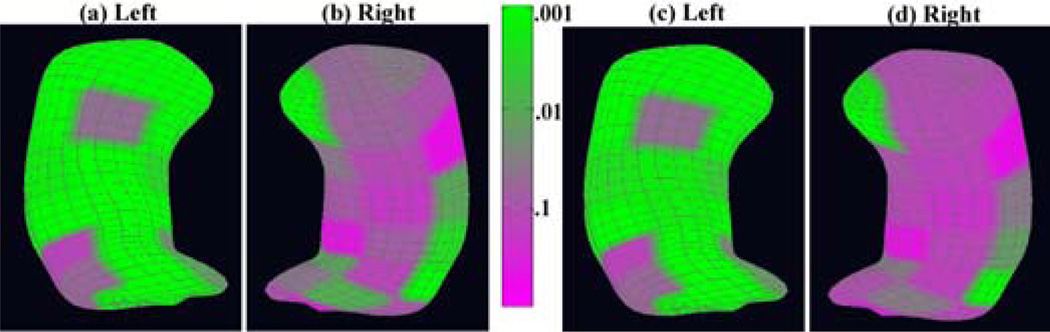
We tested the diagnostic status effect on the whole m-rep structure. We presented the color-coded p-values of the diagnostic status effects across the atoms of both the left and right reference hippocampi in Fig 2 (a) and (b) and the corresponding adjusted p-values using false discovery rate were shown in Fig 2 (c) and (d). We observed large significance area in the left hippocampus, and some in the right hippocampus even after correcting for multiple comparisons.
Fig. 2.
Results for the m-rep shape analysis result mapped to the surface of the hippocampal schizophrenia study: the color-coded uncorrected p–value maps of the diagnostic status effects for (a) the left hippocampus and (b) the right hippocampus; the corrected p–value maps for (c) the left hippocampus and (d) the right hippocampus after correcting for multiple comparisons
4 Discussion
We have developed an intrinsic regression model for the analysis of manifold-valued data as responses in a Riemannian manifold and their association with a set of covariates. We have developed an estimation procedure to calculate the intrinsic least square estimates. We have developed score statistics for testing linear hypotheses on unknown parameters. We plan to apply our method to other manifold-valued data including the Grassmann manifold, planar shapes, and deformation field.
Footnotes
This work was supported in part by NSF grants SES-06-43663 and BCS-08-26844 and NIH grants UL1-RR025747- 01, R01MH08663 and R21AG033387 to Dr. Zhu, NIH grants R01NS055754 and R01EB5-34816 to Dr. Lin, Lilly Research Laboratories, the UNC NDRC HD 03110, Eli Lilly grant F1D-MC-X252, and NIH Roadmap Grant U54 EB005149-01, NAMIC to Dr. Styner.
References
- 1.Pennec X. Intrinsic statistics on riemannian manifolds: Basic tools for geometric measurements. Journal of Mathematical Imaging and Vision. 2006;25:127–154. [Google Scholar]
- 2.Davis BC, Bullitt E, Fletcher PT, Joshi S. Population shape regression from random design data; IEEE 11th International Conference on Computer Vision; 2007. [Google Scholar]
- 3.Pennec X, Fillard P, Ayache N. A riemannian framework for tensor computing. International Journal of Computer Vision. 2006;66:41–66. [Google Scholar]
- 4.Fletcher PT, Venkatasubramanian S, Joshi S. The geometric median on riemannian manifolds with application to robust atlas estimation. NeuroImage. 2009;45:S143–S152. doi: 10.1016/j.neuroimage.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yushkevich P, Fletcher P, Joshi S, Thall A, Pizer SM. Continuous medial representations for geometric object modeling in 2d and 3d. Image and Vision Computing. 2003;21:17–28. [Google Scholar]
- 6.Jupp PE, Kent JT. Fitting smooth paths to spherical data. Applied Statistics. 1987;36:34–46. [Google Scholar]
- 7.Schwartzman A, Robert FD, Taylor JE. Cross-subject comparison of principal diffusion direction maps. Magnetic Resonance in Medicine. 2005;53:1423–1431. doi: 10.1002/mrm.20503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartzman A. Ph. D. thesis. Stanford University; 2006. Random ellipsoids and false discovery rates: Statistics for diffusion tensor imaging data. [Google Scholar]
- 9.Huckemann S, Hotz T, Munk A. Intrinsic shape analysis: Geodesic pca for riemannian manifolds modulo isometric lie group actions (with discussions) Statistica Sinica. 2009 [Google Scholar]
- 10.Miller MI, Qiu A. The emerging discipline of computational functional anatomy. NeuroImage. 2009;45:S16–S39. doi: 10.1016/j.neuroimage.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fletcher P, Joshi S, Lu C, Pizer S. Principal geodesic analysis for the study of nonlinear statistics of shape. IEEE Transactions on Medical Imaging. 2004;23:995–1005. doi: 10.1109/TMI.2004.831793. [DOI] [PubMed] [Google Scholar]
- 12.Dryden I, Mardia K. Statistical shape analysis. New York: John Wiley and Sons; [Google Scholar]
- 13.Dryden I, Mardia K. Multivariate shape analysis. Sankhya. 1993;55:460–480. [Google Scholar]
- 14.Kendall DG. Shape manifolds, procrustean metrics and complex projective spaces. Bull. Lond. Math. Soc. 1984;16:81–121. [Google Scholar]
- 15.Kent J. The complex bingham distribution and shape analysis. Journal of the Royal Statistical Society B. 1994;56:285–299. [Google Scholar]
- 16.Kim PT, Koo JY. Statistical inverse problems on manifolds. Journal of Fourier Analysis and Applications. 2005;11:639–653. [Google Scholar]
- 17.Fréchet M. Les éléments aléatoires de nature quelconque dans un espace distancé. Ann. Inst. H. Poincaré. 1948;10:215–230. [Google Scholar]
- 18.Mardia K, Jupp PE. Directional statistics. John Wiley: Academic Press; 1983. [Google Scholar]
- 19.Styner M, Lieberman JA, Pantazis D, Gerig G. Boundary and medial shape analysis of the hippocampus in schizophrenia. Medical Image Analysis. 2004;4:197–203. doi: 10.1016/j.media.2004.06.004. [DOI] [PubMed] [Google Scholar]