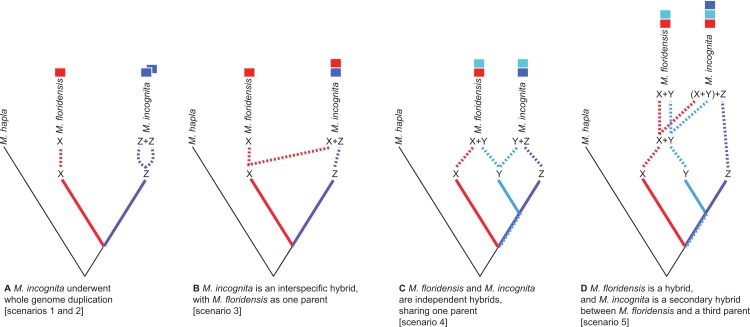
Figure 2. Scenarios of the possible relationships between Meloidogyne floridensis, Meloidogyne incognita and Meloidogyne hapla, and the origins of duplicated gene copies.
M. hapla is a diploid species in a different sub-generic group to that of M. incognita and M. floridensis. Species “X”, “Y” and “Z” are postulated ancestral parents that could have given rise to M. incognita and M. floridensis. (A) Scenarios 1 and 2: Here M. floridensis is a diploid sister species to M. incognita and possesses the “X” genome. Scenario 1 postulates reacquisition of apomixis in M. floridensis from an apomict ancestor, while Scenario 2 postulates that the apomicts repeatedly lost meiosis independently. Under both these scenarios, the presence of significant duplications in M. incognita suggests that it has undergone whole genome endoduplication. The duplicated genomes (“Z + Z”) in M. incognita are diverging under Muller’s ratchet. (B) Scenario 3: Ancestor “X” gave rise to the diploid species M. floridensis, and also interbred with “Z” to yield M. incognita, which thus carries two divergent copies of each gene (“X + Z”). In this model only M. incognita, not M. floridensis, is predicted to carry two homeologues of many genes. (C) Scenario 4: Both M. floridensis (“X + Y”) and M. incognita (“Y + Z”) are hybrid species, and share one parent (“Y”). In this model both M. incognita and M. floridensis are predicted to carry two homeologues of many genes. (D) Scenario 5: Both M. floridensis (“X + Y”) and M. incognita (“X + Y + Z”) are hybrid species, but M. incognita is a triploid hybrid between the hybrid M. floridensis ancestor (“X + Y”) and another species (“Z”). In this model M. incognita is predicted to carry three, and M. floridensis is predicted to carry two, homeologues of many genes.