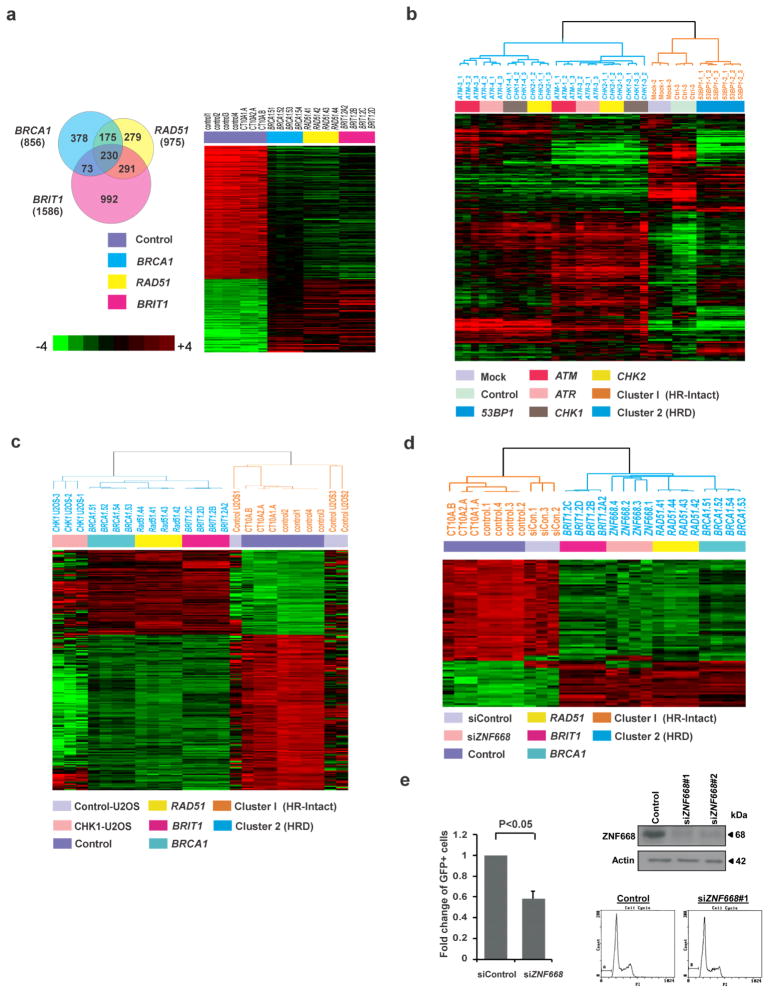
Figure 1. Gene Expression Analysis Identifies an HRD Gene Signature that Functionally Predicts the Status of HR Repair Deficiency.
(a) (Left) Venn diagram indicating numbers of genes whose expression differed between each HRD cell line and the other HRD cell lines and the control cells. The analysis was performed using BRB-ArrayTools. (Right) Heat map of the HRD gene signature, consisting of 230 genes whose expression differed between each HR-deficient cell line and the control cells. Microarray was conducted in 7 independent samples of control cells and 4 independent samples of each individual knockdown cell line. Student t-test was conducted between the average of control cells and that of knockdown cells (P<0.001).
(b) MCF-10A cells were infected by lentiviral particles targeting ATM, ATR, CHK1, CHK2, or 53BP1, and (c) U2OS cells were transfected by the ON-TARGET-plus CHK1 siRNAs. Microarray analyses were conducted to verify accuracy and specificity of the HRD gene signature by supervised clustering analysis.
(d) MCF-10A cells were transfected by the ON-TARGET-plus ZNF668 siRNAs. Microarray analysis was conducted to verify the presence of the HRD gene signature by supervised clustering analysis.
(e) U2OS cells were transfected with ZNF668 siRNA or control siRNA and analyzed for HR repair efficiency. Western blots demonstrating effective knockdown are shown to the upper right and cell cycle analysis with propidium iodide staining performed seventy-two hours after transfection are shown to the lower right.