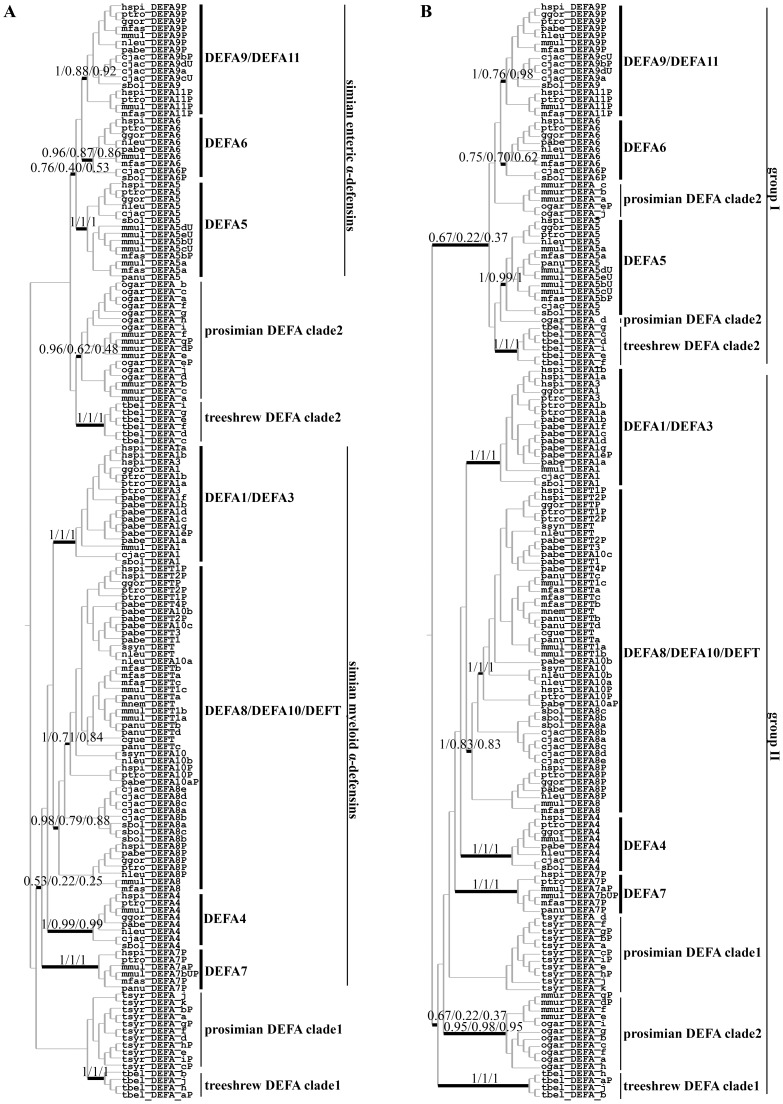
Figure 1. Phylogenetic trees of the α-/θ-defensin (DEFA/DEFT) genes in primates and treeshrews.
A: Phylogenetic tree of primate and treeshrew DEFA/DEFT genes based only on the signal-prosegment region. The BI tree is selected as the background tree. The major clades or clusters having similar topologies from all three tree-building methods (BI, NJ and ML) are combined and labeled with the BI posterior probabilities and the bootstrap support values from the NJ and ML analyses. Primate DEFA/DEFT genes are clustered into prosimian and simian clades. The simian clades are classified and named following the nomenclature used for the human DEFA/DEFT genes. The “P” in node labels denotes a pseudogene, and the “U” indicates a gene with an ambiguous locus from the species in the synteny map in Figure 7. B: Phylogenetic tree of primate and treeshrew DEFA/DEFT genes based the entire coding region. The BI tree is selected as the background tree. The treeshrew DEFA genes are the outgroups of the two separate clades.