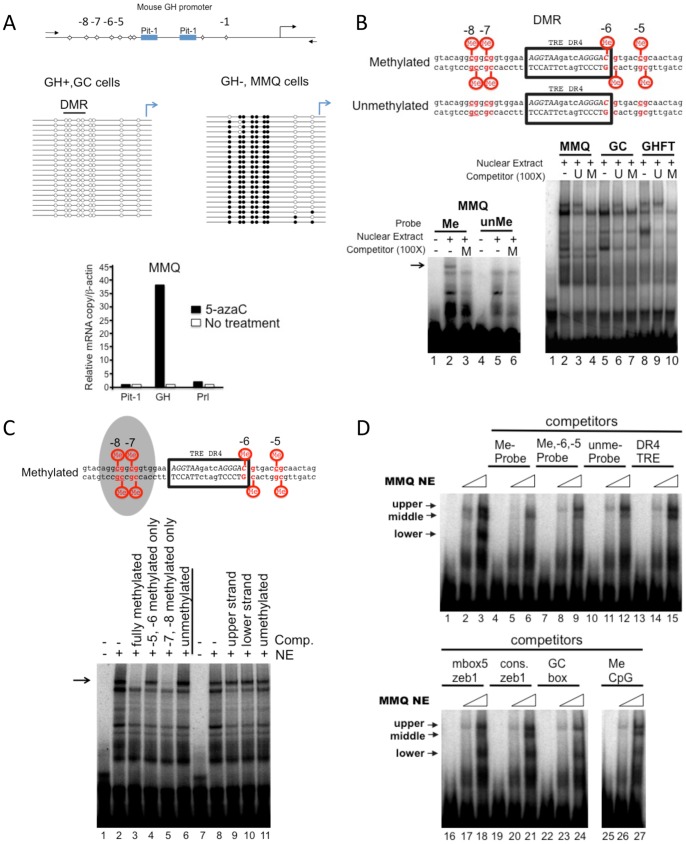
Figure 3. Inhibition of DNA methylation relieves transcriptional repression of GH and characterization of a methyl-DNA binding activity.
A. GH gene repression can be relieved by treatment with 5-azaC. Above, schematic illustration of the CpG sites location in the rat GH promoter (the CpGs are numbered according to their relative position from the transcriptional start site). Middle, bisulfite sequencing of genomic DNA extracted from pituitary derived-GH+ (GC cells) or GH− cells (MMQ cells). The level of DNA methylation is displayed as unmethylated (open circles) or methylated (solid circles) from individual clones. Lower, GH− cells (MMQ) were treated with 5-azaC and the level of gene expression compared using RT-qPCR. The solid bars represent 5-azaC treated cells and the open bars, cells cultured under normal conditions. B. A methyl-specific binding protein binds to a region of DNA derived from the mouse GH promoter. Above, DNA sequence of the double-stranded oligonucleotides used in the EMSA. The position of the CpGs relative to the transcriptional start site are indicated and the position of a previously described binding site for the thyroid hormone receptor (TR) is boxed. Lower left, EMSA with MMQ nuclear extracts comparing the bound proteins from either methylated (lane 2) or unmethylated DNA probes (lane 5). A methylated competitor DNA (M) reveals a specific upper methyl-DNA protein-binding component (Lane 3, arrow). Lower right, an EMSA competition assay with nuclear extracts from pituitary-derived cell lines (MMQ, GC and GHFT) as indicated. Methylated or unmethylated (U) competitor oligonucleotides were used to identify the upper shifted component representing a DNA bound protein. All nuclear extracts contained a methyl-DNA binding protein. C. Methylation of CpGs at position −8 and −7 were required for recruiting a methyl DNA binding protein. Above, schematic of the result from the competition assay illustrated below. The shaded area indicates CpGs essential for the methyl DNA binding protein. Below, an EMSA with MMQ nuclear extracts (NE) and a methylated DNA probe with various competitor oligonucleotides as indicated. D. The methyl-DNA binding activity observed with the GH DMR appears to be unique. The sequence of the oligonucleotides used in the competition assay are listed in Table S1. An EMSA with increasing amounts of MMQ NE (6 and 12 µg) and a methylated GH DMR probe optionally in the presence of competitor oligonucleotides (100X) as indicated.