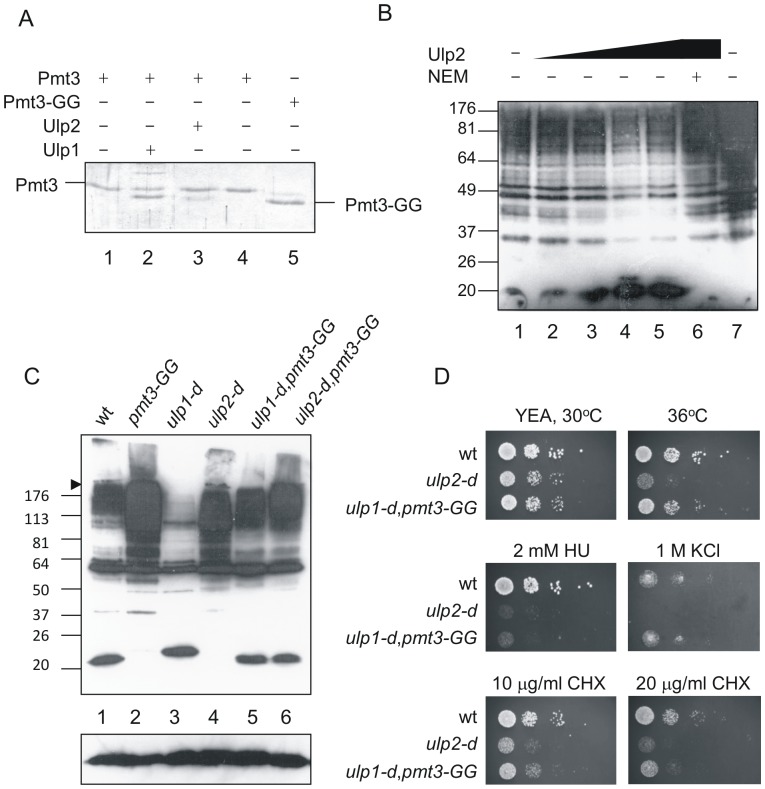
Figure 1. Analysis of Ulp2 function.
A. Assay for SUMO-processing activity. Lanes 1–4 contain full length SUMO, lane 5 SUMO-GG. Lanes 1,5, unincubated controls, lanes 2–4 were incubated at 20°C for 2 h following addition of 0.72 µg Ulp1 (lane 2), 2.32 µg Ulp2 (lane 3) or 2 µl buffer (lane 4). Proteins were analysed by SDS PAGE followed by staining with Coommassie Brilliant Blue. B. Assay for de-conjugating activity. S. pombe cell extracts were prepared using standard native extraction procedures. Extracts were incubated at 20°C for 2 h (lanes 1–6), lane 1 5 µl of fraction from extract from E. coli cells transformed with empty vector, equivalent in volume to the Ulp2-containing fraction from ulp2-transformed cells, lane 2 0.6 µg Ulp2, lane 3 1.2 µg Ulp2, lane 4 2.4 µg (5 µl) Ulp2, lane 5 4.8 µg Ulp2, lane 6 1.2 µg Ulp2 pre-incubated with 5 mM NEM, lane 7 total cell extract without incubation at 20°C. Assays were analysed by Western blotting with anti-SUMO antisera. C. Western analysis of total cell extracts using anti-SUMO antisera. Both the separating and stacking gels (6% polyacrylamide in the stacking gel) were blotted. D. Ten microlitre of 10 fold serial dilutions of cells were plated onto YEP agar plates with or without additives as indicated. 10x amount of cells of ulp2-d and ulp1-d,pmt3-GG were used compared to wild type.