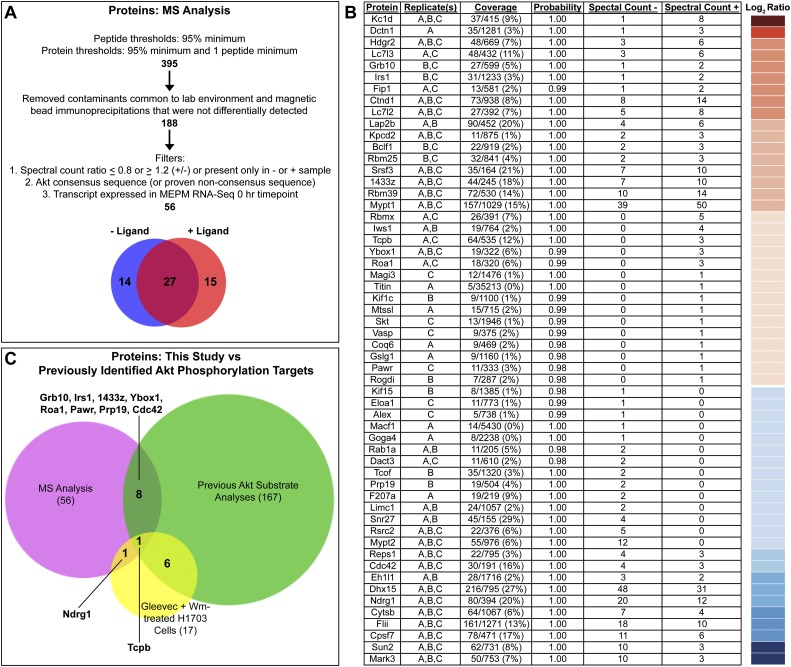
Figure 4.
Mass spectrometry-based identification of PDGF-AA-dependent Akt phosphorylation targets in primary MEPM cells. (A) Filters applied to the nano-LC-MS/MS data set. (B) Proteins identified are listed based on the ratio of the number of quantitative spectra detected in PDGF-AA-treated (+) versus untreated (−) samples. The log2 of this ratio for each protein is represented colorimetrically in a heat map at right. Additional information listed includes protein name, detection in biological replicates, coverage, protein identification probability, and quantitative spectral counts for untreated (−) and PDGF-AA-treated (+) samples as determined using Scaffold 3 software. Replicate(s), coverage, and protein identification probability pertain to the sample (untreated or PDGF-AA-treated) in which each protein had the highest number of quantitative spectra. (C) Comparison of the proteins identified in this study (56) versus previously identified Akt phosphorylation targets in humans, mice, and/or rats (167) versus proteins for which phosphorylation at an Akt consensus site is sensitive to independent treatment with the PDGFRα inhibitor Gleevec and the PI3K inhibitor Wortmannin (Wm) in H1703 cells, a non-small-cell lung cancer cell line driven by PDGFRα (17) (Moritz et al. 2010). Of the proteins identified in this study, 46 are potentially novel Akt phosphorylation targets.