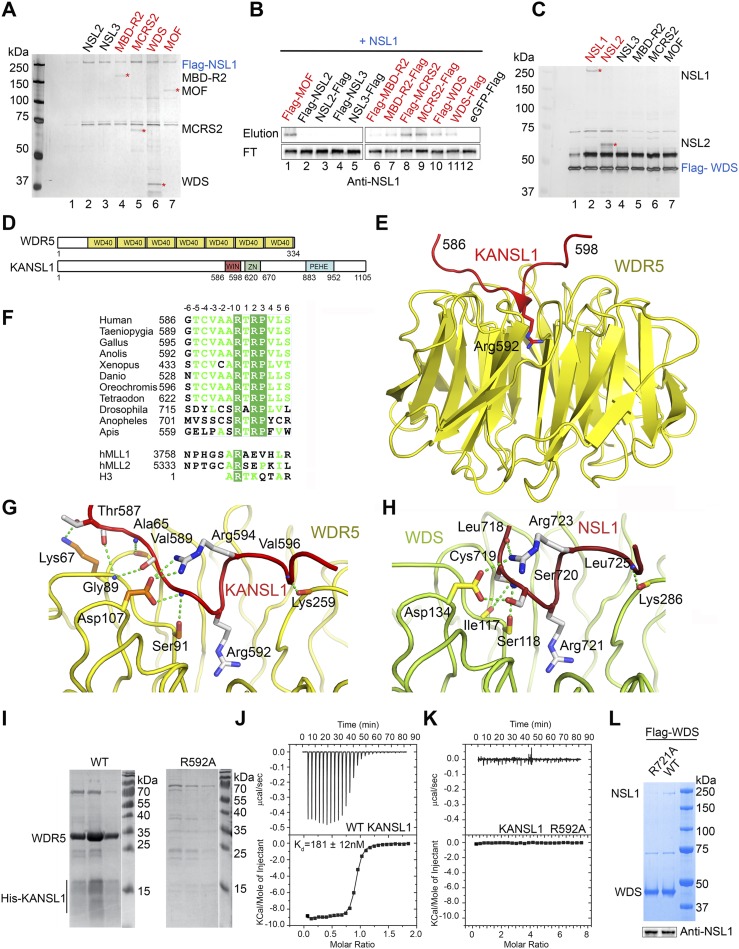
Figure 1.
Structural analysis of the KANSL1–WDR5 interaction. (A) SDS-PAGE and silver staining analysis of Flag pull-down assays using N-terminally 3xFlag-NSL1 as the bait and other untagged NSLs as prey. All proteins are from baculovirus-expressed protein extracts. Interacting proteins are indicated by asterisks and highlighted in red. (See Supplemental Fig. S1A for the input sample). (B) Western blot analysis of the reverse Flag pull-down assays of A using untagged NSL1 as the prey and either N-terminal or C-terminal 3xFlag-tagged NSLs as bait. (FT) Flowthrough sample. Interacting proteins are highlighted in red. The eluted prey proteins are shown in Supplemental Figure S1B. (C) SDS-PAGE and silver staining analysis of Flag pull-down assays using 3xFlag-WDS as the bait and other untagged NSLs as prey. All proteins are from baculovirus-expressed protein extracts. Interacting proteins are indicated by asterisks and highlighted in red. (See Supplemental Fig. S1C for the input sample). (D) Schematic representation of the domain structure of human WDR5 and KANSL1. KANSL1 contains a PEHE domain involved in the binding of MOF (Kadlec et al. 2011) and a WIN motif identified in this study. (E) Ribbon diagram of the minimal human WDR5–KANSL1 complex structure. WDR523–334 is shown in yellow, and KANSL1 is in red. The key interacting KANSL1 Arg592 is shown as sticks. (F) Sequence alignment of NSL1 proteins. Only the sequence of the fragment involved in the interaction with WDR5 is shown. Identical residues are in green boxes. Numbering of the motif residues centered on Arg592 (in humans) is shown above the alignment. Representative sequences of corresponding MLL and H3 motifs are aligned with NSL1. (G) Details of the interaction between the β-propeller domain of WDR5 and the WIN motif of KANSL1. Interacting WDR5 residues are shown in orange, and KANSL1 residues are in gray. Residues without side chains represent main chain interactions. Contacts with Arg592 of KANSL1 are similar to the ones of H3 and MLL proteins and are not shown for clarity (see Supplemental Fig. S3B). (H) Interaction interface of the complex between the β-propeller domain of WDS (green) and the WIN motif of NSL1 (red). (I) SDS-PAGE analysis of the binding of His-tagged KANSL1584–690 (wild type [WT] and the R592A mutant) to coexpressed untagged WDR523–334 after purification using Ni2+ resin. KANSL1584–690 is only detectable (but still degrading) when bound to WDR5. (J) ITC measurement of the interaction between WDR523–334 and the KANSL1 WIN motif-containing peptide (585-DGTCVAARTRPVLS-598-Y). The bottom panel represents a fit of the calorimetric data to single-site-binding model. Dissociation constant (Kd) derived from the fit is indicated. (K) ITC measurement of the interaction between WDR523–334 and the mutated KANSL1 WIN motif-containing peptide (585-DGTCVAAATRPVLS-598-Y). (L, top panel) 3xFlag-WDS was coexpressed with either wild-type NSL1 (WT) or mutant NSL1 (R721A) in a single baculovirus and purified using Flag-M2 resin. (Bottom panel) The Western blot shows that equal amounts of wild-type and mutant NSL1 (R721A) were present in the input extracts, yet only the wild-type NSL1 interaction is detected with WDS in the Coomassie blue gel.