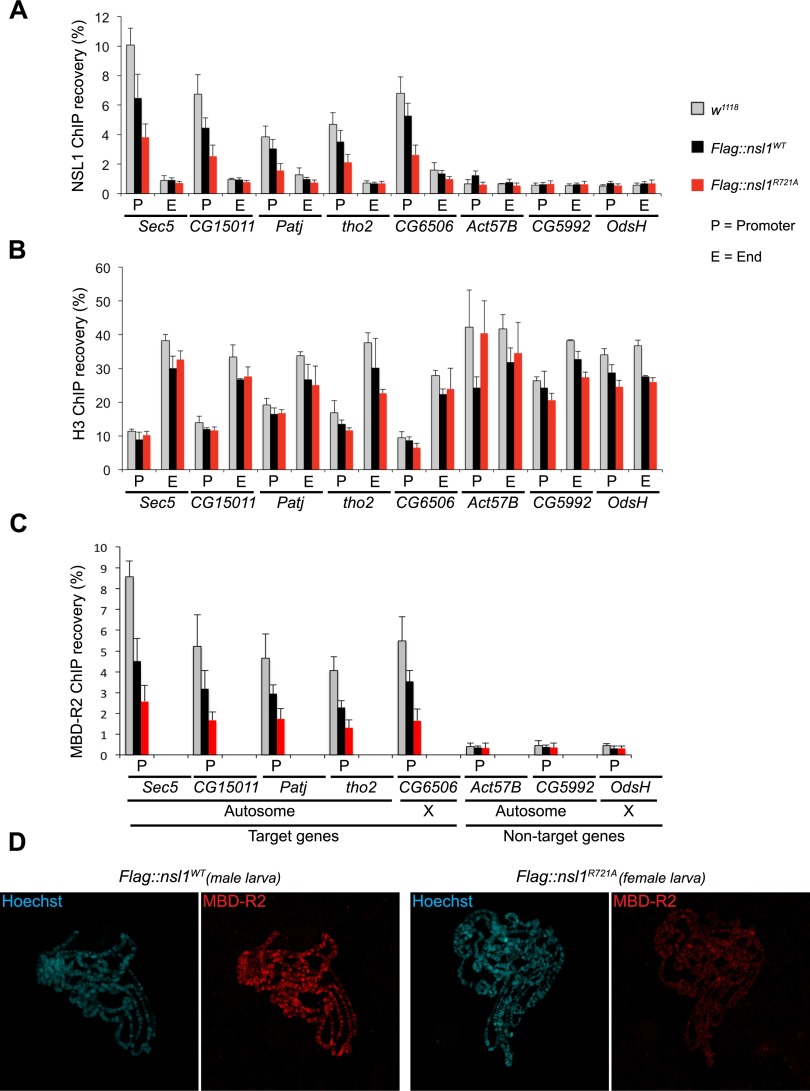
Figure 4.
The NSL1–WDS interaction is required for proper targeting of NSL1 to the promoters of target genes. (A) NSL1 ChIP from whole male larvae. qPCR was performed using primers corresponding to the promoter (P) and end (E) regions of genes. The list of target and nontarget genes was chosen based on the genome-wide data (Raja et al. 2010; Lam et al. 2012). ChIP recovery was calculated over the input DNA (shown as percentage [%]). The end of target genes and both the promoter and the end of nontarget genes were used as negative controls. The error bars represent standard deviations of three biological replicates. (B) As in A, but this time Histone H3 ChIP from whole male larvae. This provides a control for the quality of the chromatin preparations. (C) As in A, but this time MBD-R2 ChIP from whole male larvae. (D) Polytene chromosome immunostaining of Flag∷nsl1WT male and Flag∷nsl1R721A female third instar larvae. Chromosomal squashes were performed on the same slide; shown images of single nuclei were cropped from the same tile scan image. Immunofluorescence staining of MBD-R2 (red) is shown; DNA is counterstained with Hoechst (cyan).