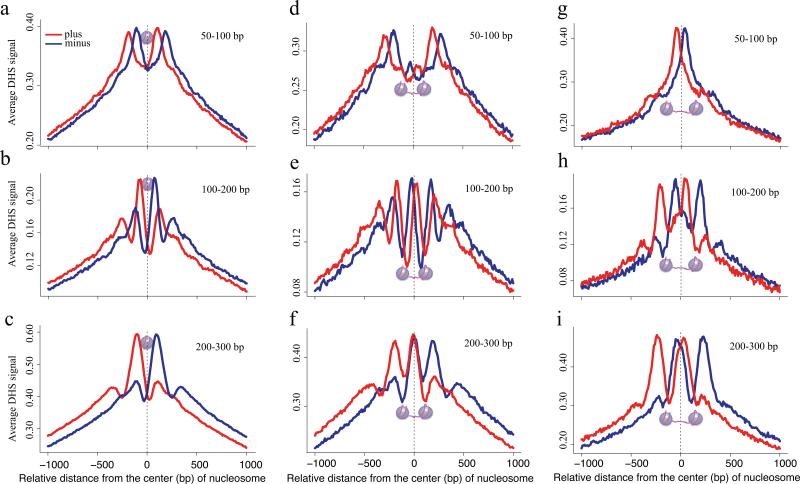
Figure 2. Nucleosome positioning effects on DNase-seq results.
(a) Schematic figure shows fragments less than 147bp in length, cannot span a nucleosome. (b, c, d) Distribution of DNaseseq tags relative to the center of nucleosomes identified by MNase digestion and H3K4me2 immunoprecipation for (b) 50-100bp, (c) 100-200bp, and (d) 200-300bp fragments in LNCaP. Tags from both the plus and minus strands for the 50-100bp fragments fall in the regions that flank the nucleosome. Plus strand and minus strand mapped ends of (c) 100-200bp and (d) 200-300bp fragments accumulate on opposite sites of the nucleosome. (e) Illustration of the 50-100bp fragments being too long to be contained entirely in the short linker (20-50bp) and too short to span the nucleosomes. (f-h) Distribution of DNase-seq tags from (f) 50-100bp, (g) 100-200bp and, (h) 200-300bp fragments relative to pairs of nucleosomes selected to have short, 20-50bp, inter-nucleosomal linker distances. The 50-100bp fragments (f) show no peak in the linker but rather show peaks on either side of the paired nucleosomes. The longer (g) 100-200bp and (h) 100-300bp fragments show peaks that are consistent with tags spanning each nucleosome in the nucleosome pair. (i-l) Nucleosome pairs with longer, 100-130bp, linkers can accommodate the (j) Short fragments entirely. (k) A minor proportion of the 100-200bp fragments can be accommodated in the linker while the majority span the nucleosome. (l) The 200-300bp fragments cannot be accommodated in the linker but they can still span the nucleosomes.