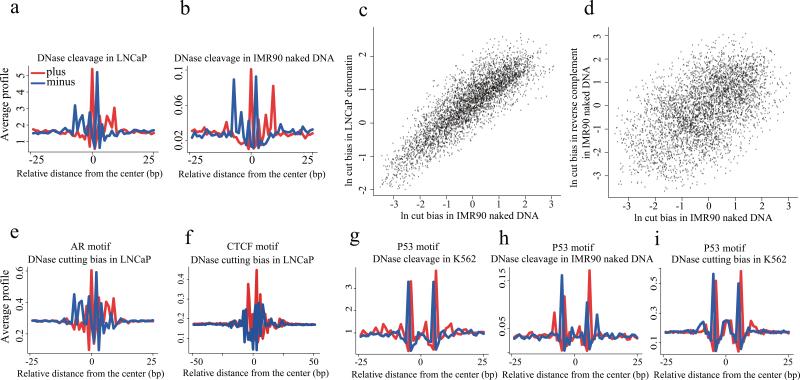
Figure 5. DNaseI cleavage bias as revealed by AR and P53 binding.
(a) The pattern of DNase cleavage across AR ChIP-seq enriched AR recognition sequences in the LNCaP cell line. (b) The DNaseI cleavage pattern produced from IMR90 naked DNA using the same AR sites as in (a). (c) The cleavage ratio represents, for each possible DNA hexamer, the number of observed cleavage sites between the 3rd and 4th bases of that hexamer relative to the number of such hexamers in the mappable genome. Cleavage ratios in IMR90 naked DNA are highly correlated with the ratios in LNCaP chromatin, showing consistency in bias across samples. (d) The log of the cleavage ratios for hexamers in DNaseI digested naked DNA and their reverse complements are plotted, showing a broad range of ratios. (e) The DNaseI cleavage pattern predicted from DNA sequence at the AR sites in (a), using the hexamer model of intrinsic DNaseI cleavage bias. (f) The pattern of cleavage predicted from a hexamer model of DNaseI cutting bias at CTCF binding sites in LNCaP. This pattern is similar to that seen in IMR90 naked DNA but different from the DNaseI cleavage pattern in chromatin at CTCF binding sites. (g) The observed DNaseI cleavage pattern in K562 chromatin at imputed p53 binding sites. (h) The DNaseI cleavage pattern produced from IMR90 naked DNA using the same p53 sites as in (g). (i) The DNaseI cleavage pattern predicted from DNA sequence using the hexamer model of intrinsic DNaseI cleavage bias at the p53 sites used in (g). Heatmaps in (a,b,e-i) show cleavage patterns at each locus for plus (red) and minus (blue) strands independently. The heatmap rows are ranked by the total DNase-seq tag count in each 50bp region.