In fission yeast, Rng8 and Rng9 form oligomers and cluster multiple Myo51 dimers to regulate Myo51 localization and function during cytokinesis.
Abstract
The myosin-V family of molecular motors is known to be under sophisticated regulation, but our knowledge of the roles and regulation of myosin-Vs in cytokinesis is limited. Here, we report that the myosin-V Myo51 affects contractile ring assembly and stability during fission yeast cytokinesis, and is regulated by two novel coiled-coil proteins, Rng8 and Rng9. Both rng8Δ and rng9Δ cells display similar defects as myo51Δ in cytokinesis. Rng8 and Rng9 are required for Myo51’s localizations to cytoplasmic puncta, actin cables, and the contractile ring. Myo51 puncta contain multiple Myo51 molecules and walk continuously on actin filaments in rng8+ cells, whereas Myo51 forms speckles containing only one dimer and does not move efficiently on actin tracks in rng8Δ. Consistently, Myo51 transports artificial cargos efficiently in vivo, and this activity is regulated by Rng8. Purified Rng8 and Rng9 form stable higher-order complexes. Collectively, we propose that Rng8 and Rng9 form oligomers and cluster multiple Myo51 dimers to regulate Myo51 localization and functions.
Introduction
Cytokinesis partitions a mother cell into two daughter cells. From yeast to humans, cytokinesis requires the coordination of four key events: cleavage site selection, actomyosin contractile ring assembly, constriction and disassembly of the contractile ring, and plasma membrane fusion and extracellular matrix formation (Balasubramanian et al., 2004; Wolfe and Gould, 2005; Barr and Gruneberg, 2007). The majority of proteins involved in cytokinesis are evolutionarily conserved (Balasubramanian et al., 2004; Glotzer, 2005; Laporte et al., 2010; Pollard and Wu, 2010). Thus, much of what we learn about cytokinesis in fission yeast is applicable to animal cells including humans.
The fission yeast Schizosaccharomyces pombe is an excellent model organism for studying cytokinesis (Gould and Simanis, 1997; Roberts-Galbraith and Gould, 2008; Lee et al., 2012). In S. pombe, cytokinesis nodes are the precursors of the contractile ring in wild type (wt) cells (Bähler et al., 1998a; Motegi et al., 2000, 2004; Paoletti and Chang, 2000; Wu et al., 2003, 2006). The nodes condense into the contractile ring through a mechanism described as the search, capture, pull, and release (SCPR) model, in which actin filaments nucleated by the formin Cdc12 from cytokinesis nodes are captured by myosin-II motors in neighboring nodes to pull them together into the contractile ring (Vavylonis et al., 2008; Coffman et al., 2009).
Myosin-Vs and its relatives are the major molecular motors that transport intracellular cargos by walking to the barbed end of actin filaments in eukaryotic cells (Sakamoto et al., 2003; Yildiz et al., 2003; Pashkova et al., 2006; Odronitz and Kollmar, 2007; Golomb et al., 2008; Hammer and Sellers, 2012; Tominaga and Nakano, 2012). These cargos include actin filaments themselves, vesicles, and various organelles (Yin et al., 2000; Mulvihill et al., 2001; Bretscher, 2003; Pruyne et al., 2004; Šamaj et al., 2005; Yu et al., 2011; Hammer and Sellers, 2012; Lo Presti et al., 2012). Myosin-Vs contain three domains: the motor head that binds ATP and actin filaments, a neck containing IQ motifs that binds light chains, and the tail (Hammer and Sellers, 2012). Their tails usually have a coiled-coil (CC) domain for dimerization and a globular tail domain (GTD) for cargo binding (Cheney et al., 1993; Pashkova et al., 2006; Eves et al., 2012). Myosin-Vs walk along actin filaments by alternating the positions of the two motor heads (Mehta et al., 1999; Sakamoto et al., 2000; Walker et al., 2000; Churchman et al., 2005; Warshaw et al., 2005). The processivity of myosin-V motors is crucial for efficient cargo transport in vivo (Rief et al., 2000; Sakamoto et al., 2003; Krendel and Mooseker, 2005; Clayton et al., 2010). Elegant in vitro and in vivo studies show that the processivity and motor activities are under sophisticated regulations by tropomyosin, adaptor proteins, cargo receptors, cargos, and cargo–actin interactions (Reck-Peterson et al., 2001; Tóth et al., 2005; Gross et al., 2007; Heuck et al., 2007; Hodges et al., 2008, 2009, 2012; Watanabe et al., 2008; Müller et al., 2009; Chung and Takizawa, 2010; Krementsova et al., 2011; Rudolf et al., 2011; Donovan and Bretscher, 2012; Sladewski et al., 2013).
Our knowledge of the roles and regulation of myosin-Vs in cytokinesis is very limited. In budding yeast, Myo2 plays a role in membrane trafficking during cytokinesis (Wagner et al., 2002; Wloka et al., 2013), but no roles in contractile ring assembly and function have been reported. Fission yeast has two myosin-Vs: Myo52 and Myo51, which have distinct localizations and functions (Mulvihill et al., 2001; Win et al., 2001; Gachet et al., 2004; Doyle et al., 2009). Their duty ratios are <50% in vitro, which indicates that they are not processive under the tested conditions (Clayton et al., 2010, 2014). However, Myo52 puncta move continuously on actin filaments in vivo (Mulvihill et al., 2006; Grallert et al., 2007; Clayton et al., 2010; Lo Presti and Martin, 2011; Lo Presti et al., 2012). Roles of Myo51 are less known. Myo51 decorates actin cables and the contractile ring, and contributes to actin cable organization (Lo Presti et al., 2012). It is also involved in cortical flow of actin filaments during cytokinesis and in the mating process (Doyle et al., 2009; Huang et al., 2012). It was unknown whether Myo51 is a functional and processive motor in vivo. Thus, the regulation of Myo51 localization and function, and how these are distinct from Myo52, remains a mystery.
Here we identify and characterize two novel CC proteins, Rng8 and Rng9, that are required for localization and function of Myo51, but not Myo52, during cytokinesis in fission yeast. Myo51 is a functional motor that can walk continuously on actin tracks in the presence of Rng8–Rng9 in vivo. It is involved in contractile ring assembly and late stages of cytokinesis together with myosin-IIs. Our data support a model in which the Rng8–Rng9 complex clusters multiple Myo51 dimers together to enable continuous movements of Myo51 on actin filaments during cytokinesis.
Results
The novel protein Rng8 localizes to the division site and is involved in cytokinesis
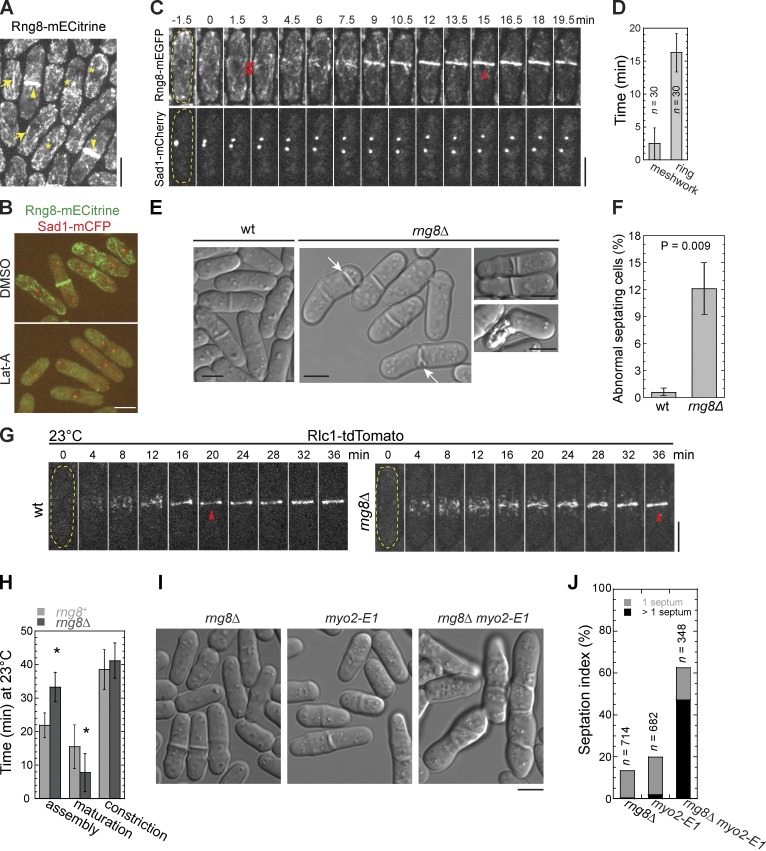
A novel protein SPAC4H3.14c was found to localize to the division site in a global analysis of protein localization in S. pombe (Matsuyama et al., 2006). It is annotated as a sequence orphan with unknown functions (PomBase No. SPAC4H3.14c). It contains three CC domains (Fig. S1 A). We further examined its localization and found that SPAC4H3.14c concentrated to cables and puncta in interphase and mainly to the contractile ring during cytokinesis (Fig. 1 A). Because of its localization to the contractile ring, we name SPAC4H3.14c hereafter as Rng8. The localizations of Rng8 depended on actin filaments because Rng8 was dispersed in cells treated with Latrunculin A (Lat-A; Fig. 1 B). Using a spindle pole body (SPB) protein Sad1 as a marker, we found that Rng8 began to form a meshwork at the cell equator 2.5 ± 2.4 min after SPB separation, which marks mitotic entry and is defined as time zero (Fig. 1, C and D; and Video 1). A compact ring of Rng8 without prominent lagging cables/strands appeared at 16.3 ± 3.0 min, and the ring constricted during septum formation (Fig. 1, C and D; and Video 1). The contractile ring localization suggests that Rng8 may have a function in cytokinesis.
Figure 1.
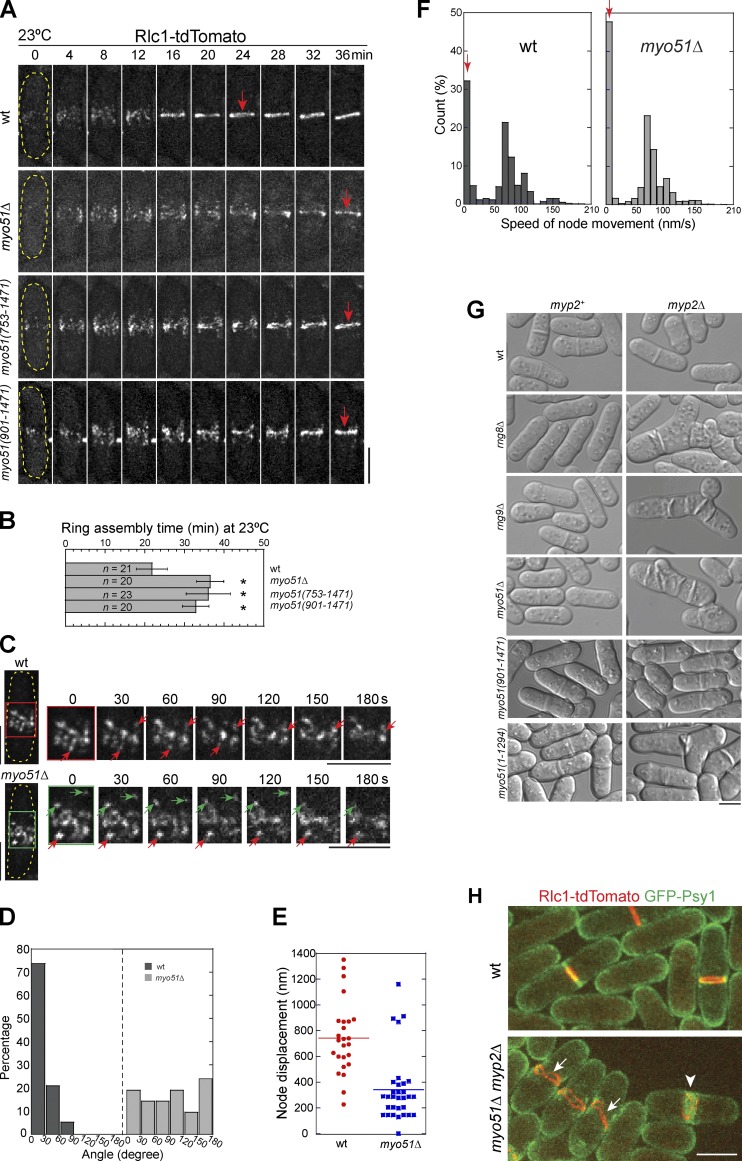
The novel protein Rng8 is involved in cytokinesis. (A) Rng8 localizes to the contractile ring (arrowheads), cables (arrows), and cytoplasmic puncta (asterisks). (B) Rng8 depends on actin filaments to localize. Cells were treated with DMSO or 100 µM Lat-A for 10 min at 25°C. (C) Time course (in minutes) of contractile ring formation in cells expressing Rng8-mEGFP Sad1-mCherry (Video 1). Time 0 marks SPB separation. The arrows and arrowhead indicate the Rng8 meshwork and the compact ring, respectively. In this and other figures, the cell boundary of some cells is marked with broken lines. (D) Time of appearance of Rng8 meshwork and the compact ring after SPB separation. Error bars indicate 1 SD. (E) Cytokinesis defects in rng8Δ cells. DIC images are shown. Arrows indicate examples of the aberrant septa. (F) Quantification of abnormal septating cells (see Materials and methods) as shown in E. n > 400 cells for each of three independent experiments. (G) Condensation of Rlc1 nodes into a compact ring (arrowheads) is delayed in rng8Δ at 23°C. Time 0 is the appearance of Rlc1 nodes. (H) Times of contractile ring assembly, maturation, and constriction in wt and rng8Δ cells (n > 25 cells for each) at 23°C. *, P < 0.001 compared with wt from two-tailed t test in this and other graphs. (I and J) Synthetic genetic interaction between rng8Δ and myo2-E1 and their septation indexes at 25°C. Bars, 5 µm.
To investigate whether Rng8 plays a role in cytokinesis, we studied the phenotype of rng8Δ cells. 12 ± 3% of septating rng8Δ cells had cytokinesis defects such as forming mislocalized, branched, or double septa and cell lysis during cell separation (Fig. 1, E and F) at 36°C. Because septation is guided by the contractile ring (Hales et al., 1999; Schmidt et al., 2002; Balasubramanian et al., 2004; Pollard and Wu, 2010), we examined contractile ring assembly (from the appearance of Rlc1 nodes to a compact ring), maturation (from a compact ring to the initiation of ring constriction), constriction, and disassembly (Wu et al., 2003) in wt and rng8Δ cells expressing myosin-II regulatory light chain Rlc1-tandem dimer Tomato (tdTomato). At 36°C, those rng8Δ cells forming branched or double septa usually had branched or double contractile rings in early cytokinesis (Fig. S1 B and Video 2), and the remaining rng8Δ cells assembled one normal ring with significant delay (Fig. S1 C). Consistently, rng8Δ cells were significantly delayed in contractile ring assembly and accelerated in ring maturation compared with wt cells at 23°C, whereas ring constriction and disassembly were similar to wt (Fig. 1, G and H). Duration of the ring maturation is shorter in most mutants that exhibit a delay in ring assembly (Coffman et al., 2009; Stark et al., 2010; Lee et al., 2012; Tebbs and Pollard, 2013); therefore, we reason that the main defect of rng8Δ lies in contractile ring assembly.
Genetic interactions of rng8Δ with other mutations further supported the finding that Rng8 functions in cytokinesis. Among the cytokinesis mutants tested, rng8Δ had strongest synthetic interactions with rlc1Δ and myo2-E1, a temperature-sensitive mutant of myosin-II heavy chain with defective motor activity (Table 1). Approximately 60–70% predicted myo2-E1 rng8Δ and rlc1Δ rng8Δ mutants failed to form colonies even at permissive temperatures. In the surviving myo2-E1 rng8Δ cells, 47% cells had >1 septum, whereas cells with multiple septa were rarely detected in the single mutants (Fig. 1, I and J). Together, these data indicate that Rng8 plays a role in cytokinesis, likely in an overlapping pathway with myosin-II.
Table 1.
Genetic interactions of rng8Δ, rng9Δ, and myo51 mutations with other cytokinesis mutations
| Parent 1a | Parent 2a | Percentage of viable double mutants at 25°C | Total number of tetrads | Genetic interaction at 25°Cb |
| rng8Δ | myo2-E1 | 33 | 19 | +++ |
| rng8Δ | rlc1Δ | 43 | 36c | +++ |
| rng8Δ | myp2Δ | 100 | 24 | ++ |
| rng8Δ | myo2-ΔIQ1 ΔIQ2 | 100 | 17 | + |
| rng8Δ | ain1Δ | 100 | 18 | +d |
| rng8Δ | cdc3-124 | 100 | 17 | +e |
| rng8Δ | cdc8-110 | 100 | 16 | − |
| rng8Δ | clp1Δ | 100 | 12 | − |
| rng8Δ | mid1-6 | 100 | 12 | − |
| rng8Δ | mid1-366 | 100 | 11 | − |
| rng9Δ | myo2-E1 | 43 | 23 | +++ |
| rng9Δ | rlc1Δ | 25 | 19 | +++ |
| rng9Δ | myp2Δ | 100 | 18 | ++ |
| myo51Δ | myo2-E1 | 43 | 17 | +++ |
| myo51Δ | rlc1Δ | 30 | 21 | +++ |
| myo51Δ | myp2Δ | 96 | 20 | ++ |
| myo51Δ | ain1Δ | 100 | 22 | +d |
| myo51Δ | rng8Δ | 100 | 17 | − |
| myo51(753–1,471) | myo2-E1 | 44 | 19 | +++ |
| myo51(901–1,471) | myo2-E1 | 45 | 22 | +++ |
| myo51(1–1,294) | myo2-E1 | 78 | 20 | ++ |
| myo51(753–1,471) | myp2Δ | 100 | 18 | − |
| myo51(901–1,471) | myp2Δ | 100 | 14 | − |
| myo51(1–1,294) | myp2Δ | 100 | 20 | ++ |
Single parent mutants were 95–100% viable on tetrad plates at 25°C.
Cells were grown exponentially in YE5S liquid medium before checking the morphology under DIC. The severity of cytokinesis defects compared to the parents (in terms of abnormal septation) was classified as: “+++,” very strong synthetic interaction and very severe cytokinesis defects; “++,” strong synthetic interaction and prominent cytokinesis defects; “+,” mild synthetic interaction and moderate cytokinesis defects; or “−,” no additive cytokinesis defects.
rng8 and rlc1 genes are tightly linked (∼34 kb) and only seven colonies were predicted double mutants in this cross.
The interaction was more obvious in EMM5S.
The interaction was more obvious at 28°C.
The novel protein Rng9 is a binding partner of Rng8
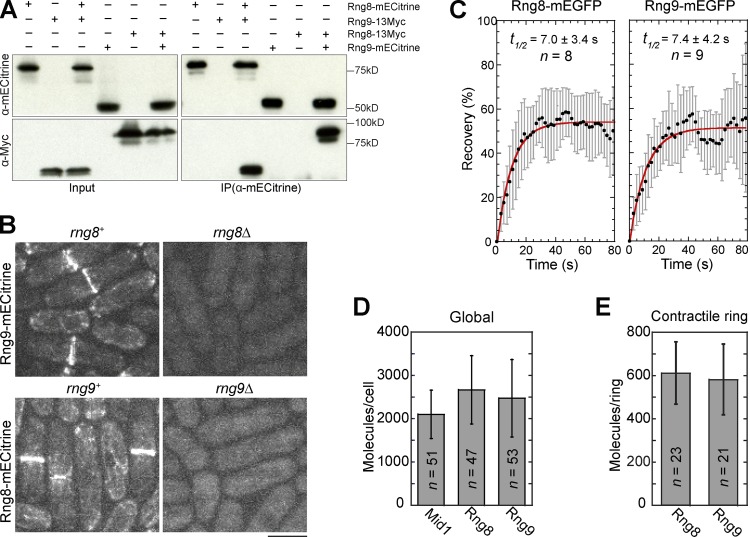
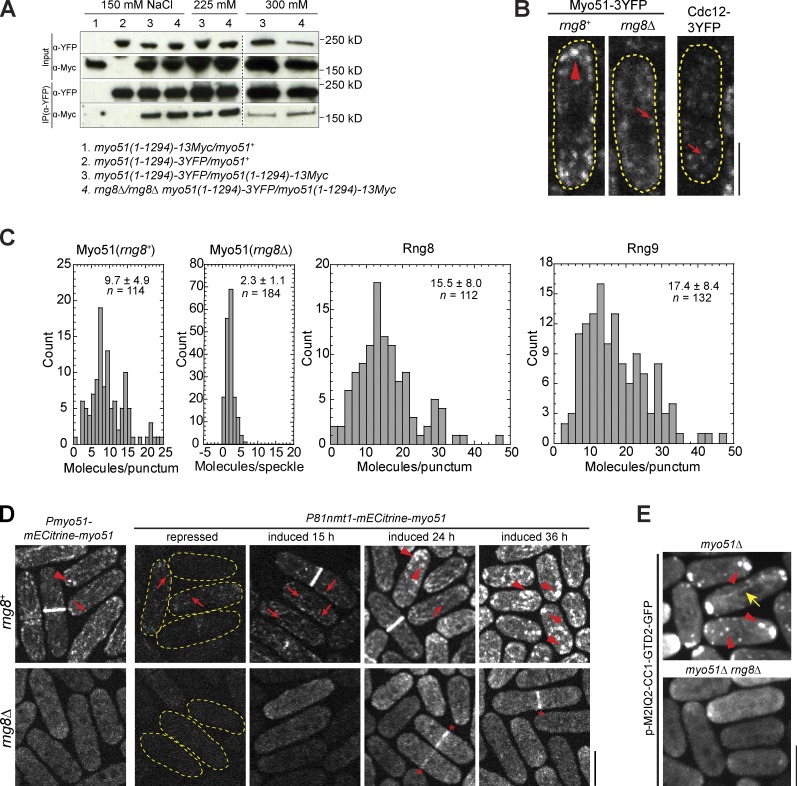
We identified another uncharacterized protein, SPBP8B7.02, as a potential binding partner of Rng8 by affinity purification of S. pombe cells expressing Rng8-S-tag and mass spectrometry (Fig. S1 D and Table S2). As presented in the following paragraph, its localization and deletion phenotype resemble those of Rng8 (Fig. 2 B and Fig. S1 F), so we named SPBP8B7.02 Rng9. Rng9 has two CC domains at its N terminus (Fig. S1 E). The physical interaction between Rng8 and Rng9 was confirmed by reciprocal coimmunoprecipitation (coIP; Fig. 2 A).
Figure 2.
Rng9 is a binding partner of Rng8. (A) Rng8 and Rng9 coIP with each other (see Materials and methods). (B) Rng8 and Rng9 are interdependent for localization. Bar, 5 µm. (C) FRAP analysis of Rng8 (left) and Rng9 (right) in the contractile ring. Curve fit is shown in red, SD in gray. (D and E) Numbers of Rng8 and Rng9 molecules globally in the whole cell (D) and locally in the contractile ring (E). Error bars indicate 1 SD.
rng9Δ had similar cytokinesis defects (Fig. S1 F) to rng8Δ (Fig. 1, E and F). In addition, like rng8Δ, rng9Δ had strong synthetic interactions with myo2-E1 and rlc1Δ (Table 1), which suggests that Rng8 and Rng9 may function in the same pathway as a complex. Indeed, Rng9 and Rng8 had identical and interdependent localizations (Fig. 2 B). However, rng9Δ cells had a slightly reduced level of Rng8, and Rng9 protein level was greatly reduced in rng8Δ cells, as revealed by Western blotting (Fig. S1 G) and fluorescence microscopy (Fig. S1 H). These data suggest that the interaction between Rng8 and Rng9 is important for both their localization and protein stability.
We further characterized Rng8 and Rng9 by studying their in vivo dynamics and stoichiometry. Both proteins were very dynamic at the contractile ring, with a recovery halftime of ∼7 s in FRAP assays (Fig. 2 C). Using Mid1-mECitrine as a known standard (Wu and Pollard, 2005), we found that each cell had 2,670 ± 790 Rng8 molecules and 2,480 ± 890 Rng9 molecules (Fig. 2 D). The compact ring had 610 ± 140 Rng8 molecules and 580 ± 160 Rng9 molecules (Fig. 2 E). These data suggest that Rng8 and Rng9 form a complex in the contractile ring with a ratio of ∼1:1.
The Rng8–Rng9 complex regulates Myo51 localization and function
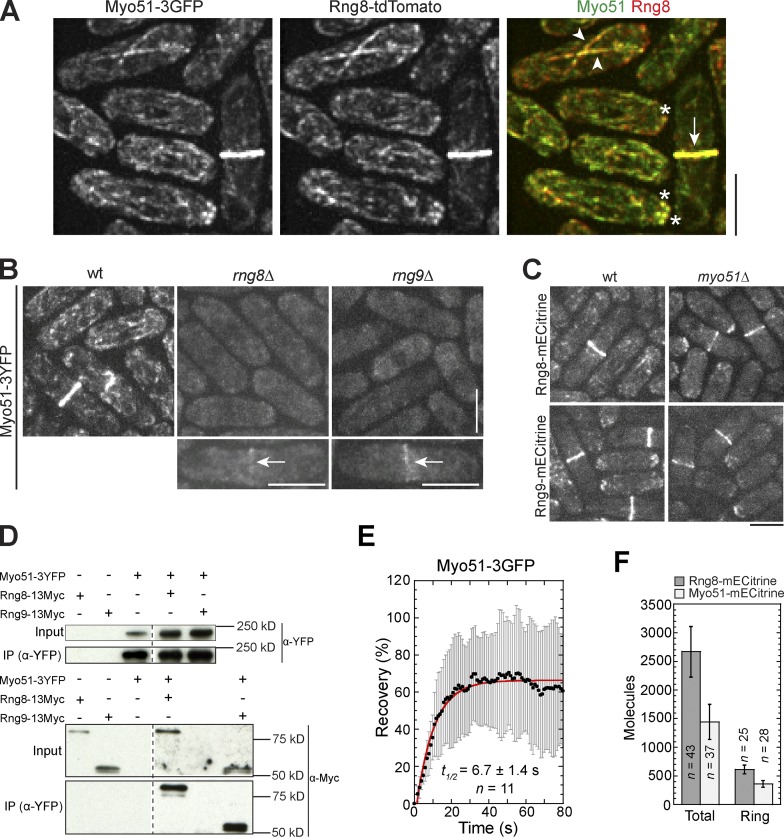
The myosin-V Myo51 also localized to the contractile ring, cables, and puncta in S. pombe (Fig. 3 A; Lo Presti et al., 2012). Interestingly, Rng8 colocalized with Myo51 in all three structures (Fig. 3 A), which suggests that the Rng8–Rng9 complex may function together with Myo51. Indeed, Myo51 localization was essentially abolished in rng8Δ or rng9Δ except for a weak signal in the contractile ring in ∼9% cells (Fig. 3 B), although the Myo51 level was not affected (Fig. S1 I). Conversely, myo51Δ only moderately reduced Rng8 and Rng9 localization to the contractile ring, while the localization to the cable was abolished (Fig. 3 C). The myosin-V Myo52 and Rng8 did not affect each other’s localization (Fig. S2, A and B). Thus, the Rng8–Rng9 complex is critical for Myo51 but not Myo52 localization.
Figure 3.
Rng8 and Rng9 are critical for Myo51 localization. (A) Colocalization of Myo51 and Rng8 in the contractile ring (arrow), cables (arrowheads), and puncta (asterisks). The imperfect colocalization on the cables is likely caused by the highly dynamic nature and movement of the structure. (B) Myo51 localization is essentially abolished without either Rng8 or Rng9. Arrows indicate weak signals left at the contractile ring in some cells. (C) Localization of Rng8 and Rng9 in myo51Δ. Bars, 5 µm. (D) Rng8 and Rng9 coIP with Myo51. Broken lines indicate that intervening lanes have been spliced out. (E) FRAP analysis of Myo51 in the contractile ring. Curve fit is shown in red, SD in gray. (F) Numbers of Myo51 molecules in the whole cells and in the contractile ring. The fluorescence intensity of Myo51 was compared with that of Rng8. Error bars indicate 1 SD.
Additional evidence further supports the finding that the Rng8–Rng9 complex functions together with Myo51. The defects of myo51Δ in cytokinesis were similar to those of rng8Δ or rng9Δ cells (Fig. S1 J). myo51Δ showed strong synthetic cytokinesis defects with myo2-E1 (Fig. S3, A–C; Huang et al., 2012) and rlc1Δ (Table 1), which suggests that the cytokinesis defects in rng8Δ and rng9Δ cells could result from the loss of Myo51 localization and function. In addition, Myo51 interacted with both Rng8 and Rng9 in coIPs (Fig. 3 D), and Myo51 was as dynamic as Rng8 and Rng9 in the contractile ring (Fig. 3 E). By comparing the fluorescence intensity, Rng8 had about twice as many molecules as Myo51 in the whole cell and in the contractile ring (Fig. 3 F). Together, these data indicate that the Rng8–Rng9 complex physically interacts with Myo51 and regulates its localization and function.
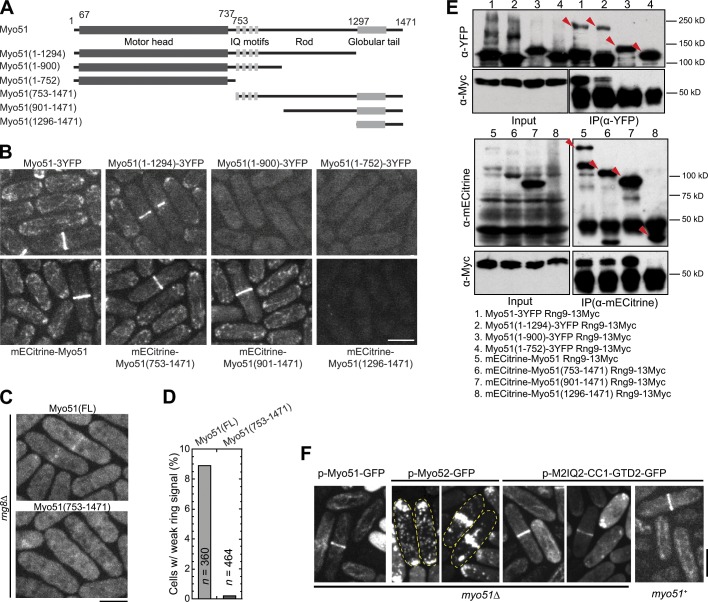
The rod region in Myo51 tail instead of its motor domain is required for Myo51 localization
To understand how Myo51 localization is regulated, we performed domain analyses for Myo51 (Fig. 4 A). Interestingly, the motor head, the IQ motifs, and GTD were not required for Myo51 localization, whereas deletion of the rod region abolished Myo51 localization (Fig. 4 B). The motor head became essential for Myo51 localization when Rng8 was absent, as full-length (FL) Myo51 but not the headless Myo51(aa 753–1,471) weakly localized to the contractile ring in rng8Δ (Fig. 4, C and D). Given that both the Rng8–Rng9 complex and the rod region of Myo51 are required for Myo51 localization, we hypothesized that the Rng8–Rng9 complex binds to the rod region. Indeed, Rng9 coimmunoprecipitated with Myo51 truncations only when the rod region was present (Fig. 4 E).
Figure 4.
The rod region in Myo51 tail determines Myo51 localization. (A) Schematics of Myo51 domains and truncations constructed. (B) Localizations of FL and truncated Myo51. (C and D) Micrographs and quantification of localization of headless Myo51(753–1,471) to the contractile ring in rng8Δ. (E) CoIP assays between Myo51 FL or truncations and Rng9. Arrowheads mark the Myo51 bands with the expected sizes. (F) The Myo51-Myo52 chimera M2IQ2-CC1-GTD2 localizes similarly to FL Myo51 in wt and myo51Δ cells. Cell boundaries are marked with broken lines. Bars, 5 µm.
To test if the rod region was sufficient for Myo51 localization and further narrow down the region responsible for localization, we constructed a chimera (M2IQ2-CC1-GTD2) in which the CC domain–containing region (aa 918–1,071) of Myo52 rod was switched to that of Myo51(903–1,078). This chimera, when expressed from a high-copy plasmid in wt or myo51Δ cells, showed similar localization to FL Myo51 but different from Myo52 (Fig. 4 F; Lo Presti et al., 2012), which indicates that aa 903–1,078 of Myo51, in the rod region of Myo51 tail, is sufficient for the localization of the myosin.
Myo51 regulates node condensation in a motor-dependent manner
To further understand the functions of Myo51 and its motor activity in cells, we next investigated the cytokinesis defects in myo51 mutants. Similar to rng8Δ cells (Fig. 1, G and H), node condensation into a compact ring took approximately twice as long in myo51Δ cells compared with wt cells, often with lagging nodes (Fig. 5, A and B; Fig. S3 D; and Video 3). In addition, the contractile ring was defective at 36°C (Video 2). The motor head of myosin-II is essential for cytokinesis in fission yeast but not in budding yeast (Lord et al., 2005; Fang et al., 2010). Cells with headless Myo51, Myo51(753–1,471), or Myo51(901–1,471) were both significantly delayed in contractile ring assembly, similar to myo51Δ cells (Fig. 5, A and B). Moreover, the two headless Myo51 had much stronger synthetic cytokinesis defects with myo2-E1 than the tailless truncation Myo51(1–1,294) (Fig. S3, A–C; and Table 1). Together, these results indicate that the motor domain of Myo51, although dispensable for Myo51 localization, is essential for its role in contractile ring assembly.
Figure 5.
Myo51 is involved in both early and late stages of cytokinesis. (A and B) Time courses (A) and quantifications (B) of contractile ring assembly in myo51Δ (Video 3) and two headless myo51 mutants using Rlc1 as the node and ring marker at 23°C. Arrows indicate the compact rings. Time 0 marks node appearance. Cell boundaries are indicated by broken lines. *, P < 0.001 compared with wt from a two-tailed t test. (C–F) Movement of Rlc1 nodes during contractile ring assembly in wt and myo51Δ at 23°C. (C) Time courses of node condensation from the boxed regions on the left. Red arrows mark nodes moving toward the center of the broad bands of nodes whereas green arrows indicate nodes moving toward cell tips for a certain time. (D–F) n = ∼30 nodes for each strain. (D) Histograms of angles of node displacements. Angles between directions of node displacements during ∼3 min and the long cell axis. (E) Node displacements during ∼3 min. (F) Distribution of instantaneous speed of node movements during ∼3 min (n > 2,000 speeds). (G) Synthetic interactions between rng8Δ, rng9Δ, or myo51 mutations and myp2Δ. Cells were grown at 36°C for 8 h before imaging. (H) myo51Δ myp2Δ cells are defective in ring stability/anchoring and disassembly during ring constriction. Arrows indicate distorted contractile ring, and the arrowhead indicates a contractile ring uncoupled from the invaginated membrane. See Video 4. Bars, 5 µm.
We next observed node movements during their coalescence into the contractile ring (Fig. 5, C–F). In wt, although nodes move in random angles in each episode of movement (Vavylonis et al., 2008), all of them moved toward the cell center and ∼70% of the nodes moved at a net angle between 0 and 30° over ∼3 min (Fig. 5, C and D). However, in myo51Δ cells, nodes moved in random directions. Only ∼50% of the nodes moved toward the cell center, and others frequently moved toward the cell tips in ∼3 min (Fig. 5, C and D), which could lead to the lagging nodes. Consistently, most nodes had shorter displacements (Fig. 5 E) in myo51Δ cells compared with wt. In addition, the absence of Myo51 led to higher frequency of nonmoving nodes (Fig. 5 F, arrows), but the moving nodes had similar instantaneous speed in wt and myo51Δ (Fig. 5 F). Thus, Myo51 regulates contractile ring formation by affecting the orientation and frequency of node movement.
Myo51 regulates late stages of cytokinesis in a motor-independent manner
rng8Δ cells are not significantly delayed in ring constriction (Fig. 1 H), and only 10–15% of myo51Δ, rng8Δ, or rng9Δ cells had abnormal septa (Fig. 1 F and Fig. S1, F and J). However, they showed strong genetic interactions (Fig. 5 G, Fig. S3 F, and Table 1) with myp2Δ, a deletion of the unconventional myosin-II that is involved in late stages of cytokinesis (Bezanilla et al., 1997, 2000; May et al., 1998; Bezanilla and Pollard, 2000; Wu et al., 2003; Samejima et al., 2010; Ye et al., 2012). The double mutants formed multiple and abnormal septa more frequently than wt and single mutants (Fig. 5 G and Fig. S3 F). These data indicate that Myo51 and the Rng8–Rng9 complex are involved in late cytokinesis in roles such as contractile ring constriction, disassembly, and/or septum formation. Surprisingly, neither of the Myo51 motor truncations had a synthetic interaction with myp2Δ. In contrast, myp2Δ was synthetic sick with Myo51(1–1,294) (Fig. 5 G, Fig. S3 F, and Table 1), although not as strong as with myo51Δ. Thus, Myo51’s role in late cytokinesis is likely motor independent.
We used Rlc1 and SNARE Psy1 (Nakamura et al., 2001) to examine the contractile ring and the plasma membrane, respectively, in myo51Δ myp2Δ mutant (Fig. 5 H and Video 4). The contractile rings were often distorted (Fig. 5 H, arrows), uncoupled from the invaginated membrane (Fig. 5 H, arrowhead), or collapsed and immediately reassembled during ring constriction (Video 4). Together, these data indicate that Myo51 also plays an overlapping role with Myp2 in late cytokinesis through a motor-independent pathway.
Myo51 affects actin cable organization and dynamics during cytokinesis
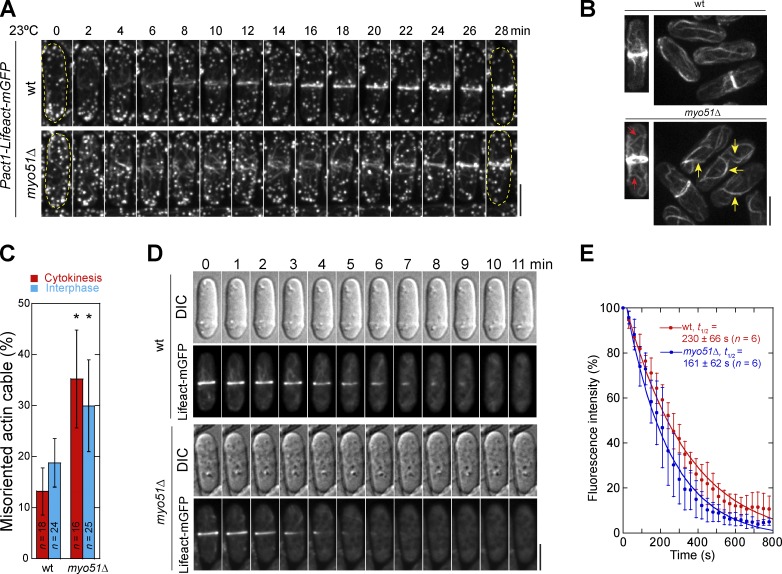
Because Myo51 is involved in actin cable organization together with Myo52 during interphase (Lo Presti et al., 2012), we checked actin structures in myo51Δ during cytokinesis. During contractile ring assembly, like actin meshwork (Vavylonis et al., 2008; Coffman et al., 2009), Myo51 meshwork mainly formed between Rlc1-labeled cytokinesis nodes (Fig. S3 E) or connected nodes (Fig. S3 E, arrow) but rarely colocalized with them, which suggests that Myo51 may affect actin filaments/bundles. Indeed, the actin meshwork at the division site, but not actin patches, was more disorganized in myo51Δ cells (Fig. 6 A and Video 5). To quantify the defects and dynamics of actin cables (Fig. 6, B and C), we treated cells with Arp2/3 inhibitor to reduce the interference of actin patches. Misoriented and curved cables increased significantly during interphase (Fig. 6 B, yellow arrows; and Fig. 6 C). During cytokinesis, not only were more actin cables detected outside the division site, but more misoriented and curved cables were also observed (Fig. 6 B, red arrows; and Fig. 6 C), which suggests that Myo51 also regulates actin cable organization besides transporting nonmedially nucleated actin filaments to the division site (Huang et al., 2012).
Figure 6.
Myo51 regulates actin structure and dynamics during cytokinesis. Actin was labeled with Lifeact-mGFP. (A) Time courses showing actin meshwork during contractile ring assembly (Video 5). Cell boundaries are indicated by broken lines. (B and C) Actin cable morphology (B) and orientation (C) during cytokinesis and interphase. Cells were treated with 100 µM Arp2/3 inhibitor CK666 for 5 min before imaging. Red and yellow arrows indicate misoriented and curved cables during cytokinesis and interphase, respectively. (C) Quantification of misoriented actin cables (see Materials and methods). Error bars indicate 1 SD. *, P < 0.001 compared with wt from a two-tailed t test. (D and E) Actin in the contractile ring is more dynamic in myo51Δ. Cells were preincubated with 100 µM CK666 for 5 min and imaged immediately after adding 4 µM Lat-A at time 0. (D and E) Micrographs (D) and fluorescence curves (E; mean ± SD) showing the fluorescence decay in the contractile ring before ring constriction. Bars, 5 µm.
We next tested whether actin stability was also affected in myo51Δ cells. The contractile ring was more dynamic in myo51Δ compared with wt (Fig. 6, D and E). Thus Myo51 affects the kinetics of actin filament turnover and stabilizes actin bundles. Consistently, myo51Δ was synthetic sick with ain1Δ (Fig. S3 G), a deletion mutant of α-actinin, which stabilizes actin filaments by cross-linking (Laporte et al., 2012). Together, our data indicate that Myo51 regulates cytokinesis through affecting actin cable organization and dynamics besides its role in cortical actin flow.
The Rng8–Rng9 complex may not be required for Myo51 dimerization
Most myosin-Vs are homodimers (Hammer and Sellers, 2012). However, Myo4 monomer in budding yeast forms a complex with the adaptor protein She3 to walk on actin (Dunn et al., 2007; Hodges et al., 2008; Heym et al., 2013). We investigated whether Rng8 and/or Rng9 affect the dimerization of Myo51 in vivo by coIP assays. Myo51-13Myc was pulled down by Myo51-3YFP in diploid cells without Rng8, Rng9, or both (Fig. S4 A), which suggests that the intermolecular interaction between Myo51 is independent of the Rng8–Rng9 complex. To rule out the possibility that Myo51 precipitates together by binding to the same cargo, we repeated the coIP using Myo51(1–1,294), the truncation lacking the cargo-binding domain (Fig. 7 A). The Myo51–Myo51 interaction was not affected by rng8Δ even at high salt concentration (Fig. 7 A). Moreover, in rng8Δ cells, Myo51-3YFP was detected in cytoplasmic speckles that had similar intensity as the formin Cdc12 speckles (Fig. 7, B and C), which are believed to be dimers (Coffman et al., 2009, 2011). On average, each speckle had 2.3 ± 1.1 Myo51 molecules in rng8Δ (Fig. 7 C). Together, these data suggest that the Rng8–Rng9 complex might not be essential for Myo51 dimerization.
Figure 7.
The Rng8–Rng9 complex promotes Myo51 clustering. (A) Myo51 self-interaction does not depend on Rng8 or Rng9 in coIP assays of cell extracts from diploid cells at different salt concentrations. Both copies of Myo51 are the truncation without the GTD domain. The broken lines indicate that intervening lanes have been spliced out. (B) Single focal plan showing Cdc12 speckles, Myo51 speckles, and Myo51 puncta imaged at the same settings. Examples of brighter puncta and dimmer speckles are marked by an arrowhead and arrows, respectively. Cell boundaries are indicated by broken lines. (C) Histograms showing the molecule numbers in Myo51 puncta (rng8+) and speckles (rng8Δ), and in Rng8 and Rng9 puncta with mean and SD indicated. The intensities were compared with those of Cdc12 speckles (see Materials and methods). (D) Myo51 punctum formation depends on Rng8. The arrowheads, arrows, and asterisks indicate the puncta, speckles, and the contractile ring, respectively. (E) Punctum formation of the Myo51-Myo52 chimera M2IQ2-CC1-GTD2 depends on Rng8. The arrowheads and arrow indicate the puncta and a cable, respectively. Bars, 5 µm.
The Rng8–Rng9 complex promotes Myo51 punctum formation
In wt interphase cells, Myo51 was frequently detected in cytoplasmic puncta (Fig. 7 B). These puncta contained more molecules than just a Myo51 dimer (Fig. 7, B and C). Only ∼30% Myo51 puncta partially overlapped with Lifeact-labeled actin patches (Fig. S4 B), which suggests that most are clusters of Myo51 molecules not bound to actin patches. Each punctum had 15.5 ± 8.0 Rng8, 17.4 ± 8.4 Rng9, and 9.7 ± 4.9 Myo51 molecules (Fig. 7 C) if we assumed each Cdc12 speckle contains two Cdc12 molecules on average (Coffman et al., 2009, 2011), which indicates a stoichiometry of ∼2:2:1 in the Rng8–Rng9–Myo51 complex. The coefficients of variation (the SD divided by the mean) are ∼50%, which suggests that the puncta tolerate some biological variations (Coffman and Wu, 2012).
We hypothesized that the Rng8–Rng9 complex promotes clustering Myo51 dimers. To test this hypothesis, we examined punctum formation with different Myo51 levels (Fig. 7 D). In rng8+ cells, when 81nmt1-mECitrine-myo51 was repressed (in YE5S) or induced for 15 h (in EMM5S), faint Myo51 speckles but not puncta were predominant (compared with wt Pmyo51-mECitrine-myo51), whereas Myo51 formed more puncta that are brighter or bigger when the cells were induced longer (24 and 36 h). This trend was not obvious in rng8Δ cells, and puncta were barely detected even after a 36-h induction (Fig. 7 D), although the overexpression did partially restore Myo51 in the contractile ring (Fig. 7 D, asterisk). Consistently, the Myo51-Myo52 chimera M2IQ2-CC1-GTD2, which mimics the localization of FL Myo51, did not localize to puncta in the rng8Δ cells (Fig. 7 E). Together, these data indicate that the Rng8–Rng9 complex clusters Myo51 dimers to promote punctum formation.
Myo51 puncta instead of speckles move continuously on actin cables in vivo
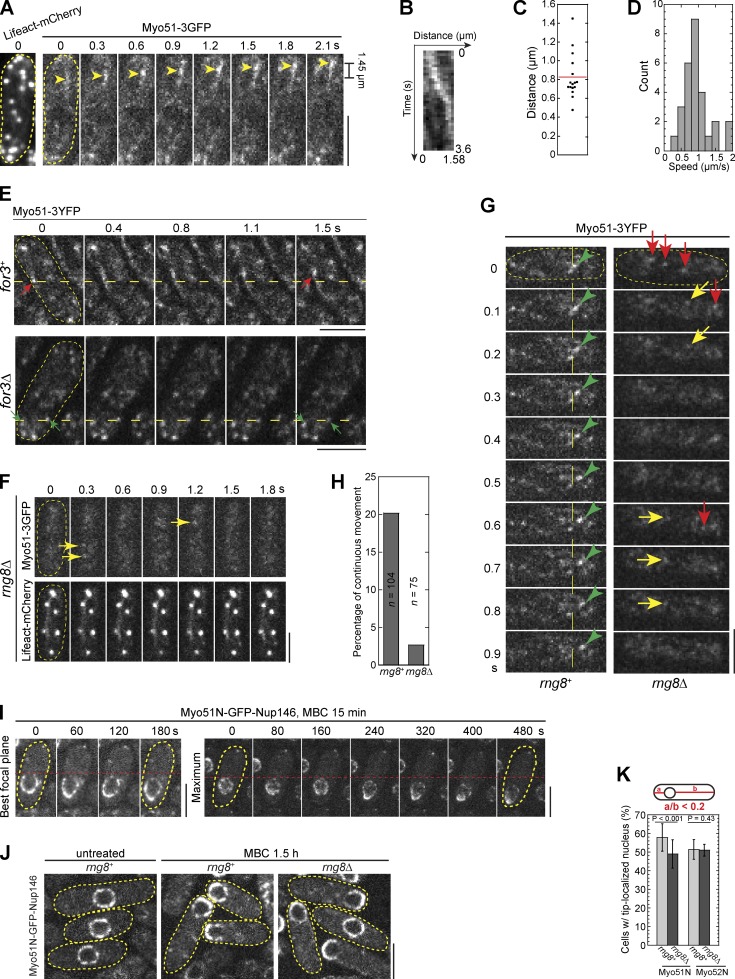
Neither Myo51 nor Myo52 are a processive motor in vitro due to their low duty ratios (Clayton et al., 2010). However, it is unclear whether they move continuously on actin cables in vivo. We tracked the movement of Myo51 on actin cables labeled with Lifeact (Huang et al., 2012). Myo51 puncta moved along actin cables continuously and unidirectionally with an instantaneous speed of 0.95 ± 0.41 µm/s and a mean travel distance of 0.83 ± 0.24 µm per episode of directional movement (Fig. 8, A–D; and Video 6). This movement is actin cable dependent because Myo51 puncta stayed still or diffused around locally in for3Δ cells where most actin cables were abolished (Fig. 8 E).
Figure 8.
Rng8 is required for continuous movement of Myo51 puncta on actin tracks. (A–D) Movements of Myo51 puncta on actin tracks in wt cells. (A) Time course of the continuous movement of a punctum (arrowheads) in a cell expressing both Lifeact-mCherry (left) and Myo51-3GFP (right). See Video 6. Cell boundaries are indicated by broken lines. (B) Kymograph showing the movement of a punctum over 3.6 s. Images were collected with 0.1-s intervals. (C) Dot plot of travel distances of Myo51 puncta. This experiment was completed once (n = 16). (D) Distribution of Myo51 instantaneous speed (µm/s). n = 28 from five puncta. (E) Myo51 punctum movement depends on actin cables. Red arrows mark a punctum moving continuously and directionally in a for3+ cell, and the green arrows mark two puncta staying still in for3Δ. Images were collected with 0.2-s intervals. (F–H) Continuous movement of Myo51 puncta depends on Rng8. (F) Time course showing that Myo51-3GFP speckles (top) in a rng8Δ cell do not move continuously on actin tracks (bottom). Arrows mark speckles that are on actin tracks at one point and disappear at the next time point. (G) Time course showing the movement of Myo51-3YFP puncta (in rng8+; green arrowheads) or Myo51 speckles (in rng8Δ). Speckles in rng8Δ either diffuse out of plane (lost track, red arrows) or diffuse around but do not move continuously (yellow arrows). See Video 7. (H) Percentage of Myo51 puncta or speckles with continuous movement, which is defined as movements that are traceable and showing unidirectional movement for ≥0.3 s in an 8-s movie with 0.1-s intervals. (I) Time courses of the nuclear displacement toward the cell tip in two cells expressing Myo51N-GFP-Nup146 after MBC treatment (Video 8). The left cell shows the best focal plane and the right shows one maximum projection. Cells were grown in EMM5S for ∼24 h and then treated with MBC for 15 min. The broken lines in E, G, and I are reference lines to aid comparisons of movements. (J) Nuclear displacement by Myo51N-GFP-Nup146 in rng8+ or rng8Δ cells 1.5 h after MBC treatment. (K) Quantification of cells with tip-localized nuclei in cells expressing Myo51N-GFP-Nup146 or Myo52N-GFP-Nup146. Definition of tip localization is shown on the top. More than 150 cells in each experiment (n ≥ 3) for each strain. Error bars indicate 1 SD. Bars, 5 µm.
In contrast, Myo51 speckles in rng8Δ cells diffused in cytoplasm instead of moving continuously on actin cables (Fig. 8, F and G; and Fig. S4 C). Consistently, ∼20% of Myo51 puncta moved continuously and directionally for a certain time during an 8-s movie in rng8+ cells, whereas Myo51 speckles had almost no directional movements in rng8Δ cells (Fig. 8, G and H; and Video 7). These data indicate that the Rng8–Rng9 complex is important for Myo51 puncta to move continuously on actin cables in vivo.
Myo51 is able to transport artificial cargo efficiently in vivo
No cargo has been identified for Myo51. To test whether Myo51 is a functional motor to transport cargos in vivo, we fused Myo51 N-terminal aa 1–1,087 (containing the motor head, IQ motifs, and the CC motifs in the rod region) to nucleoporin Nup146, the fission yeast orthologue of Saccharomyces cerevisiae Nup159 that localizes to the cytoplasmic side of the nuclear pore complex (Stelter et al., 2007; Lo Presti et al., 2012). The fusion protein mainly localized to the nuclear envelope and occasionally localized to the division site during cytokinesis (Fig. 8, I and J; Fig. S2 C; and Fig. S4 D). The division site but not nuclear localization depended on Rng8 (n > 600 cells; Fig. S2 C). In contrast, rng8Δ had no effect on the division site localization of Myo52N-GFP-Nup146 (Fig. S2 C). We tested whether the Myo51N-GFP-Nup146 was able to displace the nucleus to the cell tips like Myo52N-GFP-Nup146 (Lo Presti et al., 2012) and whether rng8Δ affected this displacement. The nucleus is normally positioned to the cell middle by microtubule pushing forces (Tran et al., 2001; Tolić-Nørrelykke et al., 2005; Daga et al., 2006). After treatment with methyl benzimidazole-2-yl carbamate (MBC) to disrupt microtubules, the nuclei of 58 ± 7% cells expressing Myo51-GFP-Nup146 moved to the cell tip (Fig. 8, I and J; and Video 8). Deletion of rng8 reduced the nuclear displacement of cells expressing Myo51N-GFP-Nup146 (49 ± 8%) but not Myo52N-GFP-Nup146 (Fig. 8 K). Consistently, Rng8 artificially colocalized with Myo51N-GFP-Nup146 but not Myo52N-GFP-Nup146 to the nuclear envelope (Fig. S4 D). Given that each nuclear pore contains multiple Nup146 (Rout et al., 2000) and a nucleus contains many nuclear pores, the artificially clustered fusion proteins around the nucleus may partially bypass the need for the Rng8–Rng9 complex–dependent clustering of Myo51. Together, these data reveal that Myo51 is a functional motor in vivo and that the Rng8–Rng9 complex is important for its transport efficiency.
Rng8 and Rng9 form preassembled stable oligomers
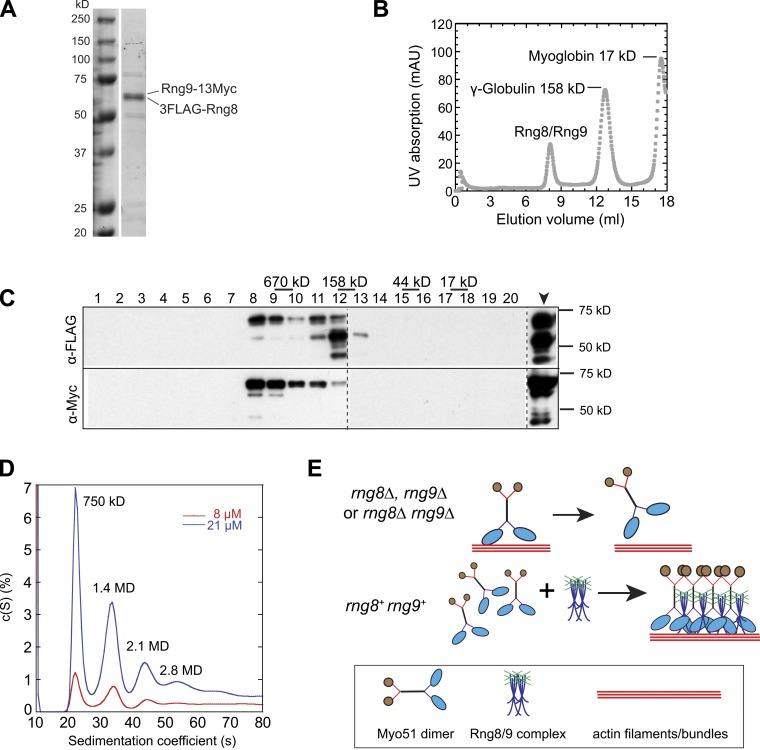
To reveal the molecular mechanism of Myo51 clustering by Rng8 and Rng9, which is important for transport and its roles in cytokinesis, we copurified both proteins from myo51Δ cells (Fig. 9 A) because myo51Δ did not abolish their interaction in vivo (Fig. S5 A). As shown by the analytical gel filtration, Rng8 and Rng9 co-migrated as a protein complex with a size >670 kD (Fig. 9, B and C; and Fig. S5 B), which suggests that Rng8 and Rng9 are capable of forming hetero-oligomers. To confirm this result and further estimate the molecular weight of the complex, sedimentation velocity analytical ultracentrifugation was performed (Fig. 9 D). Rng8 and Rng9 appeared to form several distinct higher-order oligomers with the apparent molecular weights of ∼750 kD, 1.4 MD, 2.1 MD, and 2.8 MD. The size of the most dominant species (∼750 kD) is consistent with the gel filtration data, and corresponds to seven or eight molecules of both Rng8 and Rng9 in the complex if assuming 1:1 stoichiometry (Fig. 2, D and E; and Fig. 7 C). This Rng8–Rng9 complex may serve as a basic building block to the observed larger oligomers, possibly through further self-association. Interestingly, the second species (1.4 MD) is consistent with the peak size of in vivo quantitative microscopy results (Fig. 7 C). Thus, Rng8 and Rng9 can form preassembled hetero-oligomers independent of Myo51, and such process may promote the clustering of Myo51 motors.
Figure 9.
Rng8 and Rng9 form higher-order complexes. (A) SDS-PAGE of purified 3FLAG-Rng8 and Rng9-13Myc from a myo51Δ strain. (B) Purified Rng8 and Rng9 form a large protein complex revealed by gel filtration. Elution of the purified Rng8–Rng9 complex mixed with standard proteins from a calibrated gel filtration column is shown. The peaks of two standard proteins are indicated. (C) Western blotting using anti-FLAG or anti-Myc antibodies to detect Rng8 or Rng9 from the elution fractions 1–20 (1 ml/fraction) and the sample before gel filtration (arrowhead). Broken lines indicate that intervening lanes have been spliced out. (D) Distribution of sedimentation coefficients of the Rng8–Rng9 complex at two protein concentrations, assuming a continuous c(S) distribution. Molecular weight for individual peaks are indicated. (E) Possible mechanism for the regulation of Myo51 by the Rng8–Rng9 complex in vivo. (top) Myo51 forms dimers without Rng8 and/or Rng9, and it easily detaches from actin filaments/bundles due to its low duty ratio (Clayton et al., 2010). (bottom) The Rng8–Rng9 complexes cluster multiple Myo51 dimers, and the motor heads cooperate to maintain the attachment and move continuously on actin filaments/bundles. This motor–track interaction may help to stabilize/organize actin filaments/bundles.
Discussion
In this study, we identified and characterized two novel CC proteins involved in cytokinesis: Rng8 and Rng9. They form large protein complexes to regulate the localization and function of the myosin-V Myo51. We propose a model in which the Rng8–Rng9 complex clusters Myo51 dimers to regulate Myo51’s motility and function in cytokinesis (Fig. 9 E).
Myo51 is involved in both contractile ring assembly and late stages of cytokinesis
The delay in node condensation in cells without Myo51 or Rng8 indicates that Myo51 plays a role in contractile ring assembly during cytokinesis. Recently, it was found that cortical flow of actin filaments nucleated outside the division site contribute to contractile ring assembly (Huang et al., 2012; Coffman et al., 2013), but the mechanism was still unknown. Both myosin-V and myosin-II are implicated in this process. Given Myo51’s involvement in cortical flow, one role of Myo51 in contractile ring assembly is to recruit actin filaments to the division site, which contributes to node condensation. Our findings that the nodes move less frequently and more often toward the cell tips in myo51Δ cells (Fig. 5, C–F) suggest that the cortical flow not only increases the available actin filaments for node condensation but also decreases the chance that nonmedial actin filaments counteract the condensation by pulling the nodes toward the tips. Another possible role for Myo51 in ring assembly is to stabilize the actin filaments formed among nodes to enhance the efficiency of node condensation. Our observations of two nodes connected by Myo51 filament (Fig. S3 E) and the changes in actin organization/dynamics in myo51Δ cells (Fig. 6) support this possibility.
Myo51 also plays a role in contractile ring constriction and stability together with the myosin-II Myp2 in late cytokinesis (Fig. 5, G and H; and Fig. S3 F). However, this function does not require the motor activity of Myo51, implying that its tail must contribute to cytokinesis. Budding yeast myosin-II Myo1 maintains most of its cellular function without a motor head (Lord et al., 2005; Fang et al., 2010; Wloka et al., 2013). It will be interesting to study whether Myo51 functions similarly in late cytokinesis as budding yeast Myo1. Because rng8Δ and rng9Δ displayed a similar additive effect with myp2Δ (Fig. 5 G and Fig. S3 F), it is quite likely that Myo51 function in late cytokinesis also requires Rng8 and Rng9.
Rng8 and Rng9 are important for the localization and function of Myo51
We showed that Myo51 puncta walk continuously on actin cables and that Myo51 displaces the nucleus efficiently when artificially fused with a nucleoporin (Fig. 8), which indicates that Myo51 is a functional motor for cargo transport in vivo. However, it was suggested that Myo51 is not processive in vitro due to its low duty ratio (Clayton et al., 2010). Our findings on Rng8 and Rng9 make it possible to bridge this discrepancy and support the model in which the Rng8–Rng9 complex clusters Myo51 dimers and coordinates their continuous movements on actin tracks (Fig. 9 E). In turn, these movements may stiffen actin filaments/cables as proposed for Myo52 (Lo Presti et al., 2012).
Two lines of evidence suggest that the Rng8–Rng9 complex may be primarily involved in clustering of Myo51 motors rather than directly recruiting Myo51 to actin cables. First, Rng8 and Rng9 are important for Myo51 localization, but a small fraction of Myo51 still localizes to the ring in rng8Δ or rng9Δ (Fig. 3 B) in a motor-dependent manner (Fig. 4, C and D), and the ring localization in rng8Δ is enhanced when Myo51 is overexpressed (Fig. 7 D). Second, rng8Δ only partially compromises the efficiency of nuclear displacement by Myo51N-GFP-Nup146 (Fig. 8, I–K), although Myo51 localization to actin cables appears essentially abolished in rng8Δ. In this case, the nucleoporin Nup146 probably artificially clustered the fusion proteins around the nucleus and bypassed the need for Rng8–Rng9 complex–dependent clustering. This experiment thus likely underestimates the effect of Rng8 on Myo51 transport of the artificial cargo. In addition, the continuous movement of the nucleus (Fig. 8, I–K), together with the unidirectional and continuous movements of Myo51 puncta but not speckles (Fig. 8, A–H), indicates the importance of Myo51 clustering to the motor movement in vivo.
The natural cargos transported by Myo51 are unknown. Given that Rng8 and Rng9 still localize to the contractile ring and the puncta at cell tips in myo51Δ cells (Fig. 3 C), it is unlikely that they are cargos of Myo51. Furthermore, the cargo-binding GTD is not required for Myo51 localization (Fig. 4 B), which suggests that the Rng8–Rng9 complex regulates Myo51 localization and function independently of cargos. However, we cannot rule out that other regions of Myo51 may bind to cargos. Loss of GTD slightly reduced Myo51 localization (Fig. 4 B), which suggests that Myo51 is also regulated by cargos. Hence, uncovering Myo51’s cargos is a priority for future research.
Comparison of myosin-Vs in fission and budding yeast
In budding yeast, Myo4-dependent transport of mRNAs to the bud tip requires two adaptor proteins: She2 and She3 (Kruse et al., 2002). The CC domain and C-terminal tail of Myo4 synergistically bind She3 rather than another Myo4 molecule (Heuck et al., 2007; Hodges et al., 2008), although there is discrepancy on the stoichiometry between She3 and Myo4 (Dunn et al., 2007; Hodges et al., 2008; Heym et al., 2013). In turn, She2 forms tetramers that bind and bring together two copies of the Myo4–She3 complex (Krementsova et al., 2011; Heym et al., 2013; Sladewski et al., 2013). The Rng8–Rng9 complex likely regulates Myo51 with a mechanism somehow different from She2 and She3 because: (1) the 2:2:1 stoichiometry of Rng8–Rng9–Myo51 in puncta (Fig. 7 C) is different from the 2:1:1 of She2–She3–Myo4 complex according to most studies; (2) Myo51 self-interacts and still localizes in the cytoplasm as a dimer without Rng8 and Rng9 (Fig. 7, A–C), which suggests that the Rng8–Rng9 complex does not affect Myo51 dimerization; and (3) Rng8 and Rng9 form oligomers in the absence of Myo51, which indicates that they are preassembled to cluster Myo51. However, the assembly of She2/She3/Myo4 is most likely cargo dependent, and requires the existence of Myo4 (Kruse et al., 2002; Krementsova et al., 2011; Heym et al., 2013; Sladewski et al., 2013).
It is interesting that the two myosin-Vs—Myo51 and Myo52—in fission yeast have distinct localizations. Mounting examples have shown that cargos provide the specification of localizations in different organisms (Kruse et al., 2002; Bretscher, 2003; Golomb et al., 2008; Donovan and Bretscher, 2012; Hammer and Sellers, 2012). We showed that the localization of Myo51, but not Myo52, largely depends on the binding of the Rng8–Rng9 complex to the rod region of Myo51. Similarly, Myo52’s rod region, but not its GTD and cargo Ypt3, is required for Myo52 localization at cell tips (Mulvihill et al., 2006; Grallert et al., 2007; Lo Presti et al., 2012; Clayton et al., 2014). However, it is unknown whether this occurs through regulating dimerization or binding to some regulators. Remarkably, replacing the rod region of Myo52 with that of Myo51 resulted in a chimeric protein localizing like Myo51. Thus, in fission yeast, the critical determinants of myosin-V localization are their rod regions.
In summary, here we identified and characterized two novel proteins Rng8 and Rng9 and uncovered their roles in regulating localization and function of the myosin-V Myo51 during cytokinesis. Our findings provide new insights into the regulation and function of myosin-Vs and their cooperation with myosin-IIs. It will be interesting to know whether myosin-Vs are also involved in cytokinesis, especially contractile ring assembly in other organisms.
Materials and methods
Strains, growth conditions, and genetic and cellular methods
Table S1 lists the S. pombe strains used in this study. Standard genetic and PCR-based gene targeting methods were used to transform yeast cells and construct strains (Moreno et al., 1991; Bähler et al., 1998b). All tagged and truncated genes are under the control of the endogenous promoters and integrated at their native chromosomal loci to replace the native genes except where noted. The N-terminal tagging and truncations of myo51 were made by first cloning the promoters of myo51 (−289 to +6) into the pFA6a-kanMX6-P3nmt1-mECitrine vector to replace the 3nmt1 promoter to obtain plasmids JQW674. This plasmid was used as a template to amplify kanMX6-Pmyo51-mECitrine fragments flanked with homologous sequences from the designated positions at the myo51 locus, which were then transformed into wt strain as described previously (Bähler et al., 1998b). The positive transformants were confirmed by PCR. The nmt1 promoters were repressed in YE5S medium or EMM5S with 5 µg/ml thiamine and induced in EMM5S. To test the functionalities of tagged FL Rng8, Rng9, and Myo51, both N- and C-terminally tagged strains were crossed to myp2Δ. The growth and morphology of double mutants were similar to a myp2Δ single mutant at different temperatures, which indicates that these tagged proteins are fully functional under the tested conditions.
The chimera M2IQ2-CC1-GTD2 was cloned under the control of the intermediate nmt1 promoter in pREP41 plasmid vector and encodes, in this order, aa 1–917 of Myo52, aa 903–1,078 of Myo51, and aa 1,072–1,516 of Myo52 and GFP. This chimera and Myo51 and Myo52 FL controls were all expressed on pREP41 plasmids in either wt or myo51Δ strains.
The chimera Myo51N-GFP-Nup146 was cloned under the control of the weak nmt1 promoter in pRIP82 plasmid vector and encodes, in this order, aa 1–1,087 of Myo51, an SGRA linker, GFP, a GSSGP linker, and FL Nup146 (UniProtKB accession No. Q09847). The cassette was then linearized and integrated into the ura4 genomic locus. The chimera Myo52N-GFP-Nup146, which was constructed as described previously (Lo Presti et al., 2012), was under the control of the weak nmt1 promoter in pRIP82 vector and encodes, in this order, aa 1–1,162 of Myo52, an SGRA linker, GFP, a GSSGP linker, and FL Nup146.
Cells were grown in an exponential phase at 25°C for 36–48 h before microscopy, as described previously (Wu et al., 2006). For quantifying cytokinesis defects of myo51Δ, rng8Δ, and rng9Δ strains, or the synthetic interactions with myp2Δ mutants at 36°C, cells were first grown at 25°C for ∼36 h, then shifted to 36°C for 8 h before imaging. Abnormal septating cells were defined as cells with: (1) >1 septa, (2) branched septa, (3) one septum outside the central 1/5 of the cell, (4) one septum not within 90 ± 10° relative to the long axis of the cell, or (5) septating cells with a lysed daughter cell. To reveal ring assembly defects of myo51Δ and rng8Δ strains at 36°C, cells were first grown at 25°C for ∼36 h, then shifted to 36°C for 4 h before imaging at 36°C. To test whether localization of Rng8 depends on F-actin, cells were treated with Lat-A at a final concentration of 100 µM from a 20-mM stock solution in DMSO (Wu et al., 2001) and imaged on bare slides to maintain the Lat-A concentration. To study actin dynamics, we treated cells with a final concentration of 4 µM Lat-A. MBC was used at a final concentration of 25 µg/ml and imaging was done on a gelatin slide with the same drug concentration (Wu et al., 2011). The Arp2/3 inhibitor CK666 (ChemDiv; Nolen et al., 2009) was used at a final concentration of 100 µM from a 10-mM stock solution in DMSO (Laporte et al., 2012). For septum staining, a final concentration of 10 µg/ml Calcofluor was used and cells were stained in the dark for 5 min before imaging (Wu et al., 2001, 2011).
Microscopy and data analysis
Microscopy was performed as described previously (Wu et al., 2006; Coffman et al., 2009). In brief, cells were collected from liquid cultures by centrifugation at 5,000 rpm, and washed twice with EMM5S to reduce autofluorescence for fluorescence microscopy. Live-cell microscopy was performed using a thin layer of EMM5S liquid medium with 20% gelatin (Sigma-Aldrich) and 50 nM n-propyl-gallate and observed at 23–25°C except where noted.
Three microscope systems were used to collect images with 100×/1.4 NA Plan-Apochromat objective lenses (Nikon): (1) a spinning disk confocal system (UltraVIEW ERS; PerkinElmer) with 440, 568-nm solid state lasers and 488, 514-nm argon ion lasers, and a cooled charge-coupled (CCD) device camera (ORCA-AG; Hamamatsu Photonics) on a microscope (Eclipse TE2000-U; Nikon) with 2 × 2 binning; (2) a spinning disk confocal system (UltraVIEW Vox CSUX1 system; PerkinElmer) with 440-, 488-, 515-, and 561-nm solid state lasers and dual back-thinned EM CCD cameras (C9100-13; Hamamatsu Photonics) on a microscope (Ti-E; Nikon) without binning, with the camera mode (single or dual) dependent on the experiments; and (3) an inverted microscope (Eclipse Ti; Nikon) equipped with a cooled digital camera (DS-Ql1; Nikon) for Calcofluor staining and some differential-interference contrast (DIC) images. Under our imaging conditions, no bleed-through between 514/515- and 561/568-nm channels was detected.
Image analyses were performed using ImageJ (National Institutes of Health), UltraVIEW, and Volocity (PerkinElmer). Images in figures were maximum-intensity projections of z sections spaced at 0.3–0.5 µm except where noted. To quantify nuclear displacement in cells expressing Myo51N-GFP-Nup146 or Myo52N-GFP-Nup146 fusion proteins, the distances from nuclear envelope to both cell tips as indicated in Fig. 8 K were measured in ImageJ. The nucleus was considered to be tip localized if the ratio of the shorter distance to the longer distance was <0.2. To simplify the quantification, only interphase cells with one nucleus were measured. We used Volocity to deconvolve images in Figs. 3 A, 7 D, and S3 E. Calculated point spread function (PSF) for the 100× objective lens was used to deconvolve the images with five iterations and 98% confidence. Error bars in all the figures indicate 1 SD.
To analyze node condensation and displacement, the top of the cell (five planes) with 0.3 µm spacing was imaged and projected to the same focal plane. Individual node movements were tracked in movies (2-s delay) using the plug-in MTrackJ in ImageJ (Laporte et al., 2012). Node displacement is the straight line distance between node positions at the beginning and end of an ∼3-min movie. Instantaneous node speed is the actual distance travelled by a node per second for each frame. The angle of displacement is the angle from the line formed by initial and end positions of the node to the long cell axis.
To compare actin cable orientations, cells were preincubated with 100 µM Arp2/3 inhibitor CK-666 for 5 min to reduce the interference of actin patches for analysis, and imaged on bare slides with 13 z sections spaced at 0.4 µm. To compare actin cable orientations during cytokinesis, cells with a compact ring were used and cables outside the ring were measured on their best focal planes. To compare actin cables in interphase cells, all visible cables were included. Misoriented cables were defined as a cable with an angle to the long axis of the cell >45°.
To analyze actin dynamics, experiments were performed as described previously (Laporte et al., 2012), with minor modifications. Cells were washed in EMM5S with 50 nM n-propyl-gallate and preincubated with 100 µM CK-666 for 5 min. Then 4 µM Lat-A was added, and cells were imaged immediately on bare slides. Here time 0 is the start of the observation. Intensities in 16 z sections spaced at 0.4 µm for each time point were summed. After subtracting background and correcting for photobleaching during image acquisition, intensity values of Lifeact-mGFP at the division site were normalized against the fluorescence intensity at time 0, which is set to 100%. Curve fits were obtained from the mean of all cells. SDs were from individual measurements. The single exponential decay curve equation is y = m1 – m2 exp(–m3 x), where m3 is the off rate. To calculate the t1/2 of the decay, we used the equation t1/2 = ln2/m3. To get a plateau for each decay curve, images were collected in 30-s intervals over 25–30 min.
FRAP analysis
FRAP experiments were performed with the Photokinesis unit on the UltraVIEW ERS confocal system as described previously (Coffman et al., 2009; Laporte et al., 2011). The full-size nonconstricting ring was selected and the middle slice from the z stacks was used for bleaching. After collecting four prebleach images, a selected region of interest (ROI) was bleached to >50% of the original signal, and ≥100 postbleach images with 1- or 2-s delay were collected. After subtracting the background and correcting for the photobleaching during image acquisition (using intensity of unbleached cells), the ROI intensity was normalized with the mean prebleach intensity set to 100% and the intensity just after bleaching set at 0%. Time 0 indicates the ending time of the bleaching. Intensity of every three consecutive time points was averaged to reduce noise. The exponential equation y = m1 + m2 exp(−m3 x) was used for the curve fit of fluorescence recovery, in which m3 is the off-rate (KaleidaGraph; Synergy Software). The half-time of recovery was calculated as t1/2 = ln2/m3.
Counting numbers of protein molecules
We counted protein molecules globally and locally using fluorescence intensity (Wu and Pollard, 2005; Wu et al., 2008; Coffman and Wu, 2012). In this paper, we define one polypeptide as one molecule. To count the total molecules per cell and the molecules in the contractile ring for Rng8, Rng9, and Myo51, we summed the intensity from 11 z sections spaced at 0.4 µm, which is close to the full width at half maximum of our confocal system (Coffman et al., 2011). The uneven illumination was corrected using images of purified 6His-mYFP solution (Coffman et al., 2009; Coffman and Wu, 2012). To obtain fluorescence intensity for each strain, 40–60 tagged cells and 20–30 wt cells were measured. The mean intensity of wt cells was used for autofluorescence correction. The intensities of mECitrine-tagged Rng8, Rng9, and Myo51 were normalized to that of Mid1-mECitrine to obtain the molecule numbers (Wu and Pollard, 2005; Laporte et al., 2011).
The mean intensities of the contractile rings were measured using a rectangular ROI that is big enough to include >90% of ring signal. The intensities of the cytoplasmic signal in a concentric rectangular ROI (approximately twice as big as the ROI that elongated along the cell long axis) were used for background subtraction (Wu and Pollard, 2005; Wu et al., 2008). We normalized the mean ring intensity to the mean total intensity of each protein in the whole cells with area correction to obtain the molecule numbers of Rng8, Rng9, and Myo51 in the contractile ring.
To count molecules in the puncta for 3YFP-tagged Rng8, Rng9, and Myo51, three z sections that contained the signal for each punctum were measured. A 5 × 5 pixel ROI was used to measure the intensity. The background intensity from a concentric 7 × 7 pixel ROI was calculated and subtracted from the measuring area in each imaging plane as described previously (Wu and Pollard, 2005; Wu et al., 2008). Then the sum intensity of the three z sections was normalized to obtain the molecule numbers per punctum based on the mean intensity of Cdc12-3YFP speckles, which were measured in the same way and were set as two molecules per speckle (Coffman et al., 2009, 2011).
Immunoprecipitation (IP) and Western blotting
We performed IP using S. pombe cell lysates according to the protocol that was described previously (Laporte et al., 2011; Lee and Wu, 2012). In brief, we washed 30 µl of protein G covalently coupled magnetic Dynabeads (Invitrogen) three times with cold PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4) and then resuspended them in 600 µl PBS. The beads were incubated with 5 µg YFP/mECitrine antibodies (NB600-308; Novus Biologicals) for 1 h at 23°C. The antibody-coupled magnetic beads were then washed with PBS and with 1% IP buffer (1% NP-40 buffer [Laporte et al., 2011], 1 mM PMSF, and protease inhibitors [Roche]). 30 mg of lyophilized cells per sample were resuspended in 300 µl of IP buffer and cleared by two rounds of centrifugation at 4°C with speeds of 3,000 rpm and 13,000 rpm, respectively. In the case of diploid cells, 40 mg of lyophilized cells were used per sample. The supernatant was added to the antibody-coupled beads and incubated for 90 min at 4°C. Beads were washed five times with cold 1% NP-40 buffer. To elute the proteins, 50 µl of sample buffer was added to the beads and boiled for 5 min.
Western blotting was performed as described previously (Laporte et al., 2011; Lee and Wu, 2012) with the following modifications: (1) A monoclonal antibody against GFP (1:2,000 dilution; catalogue No. 11814460001; Roche) was used to detect Rng8-mECitrine, Rng9-mECitrine, and Myo51-3YFP; and (2) a monoclonal antibody against FLAG (1:2,000 dilution; catalogue No. F1804; Sigma-Aldrich) was used to detect 3FLAG-Rng8.
To compare protein levels for Rng8, Rng9, and Myo51, 100 µl of IP buffer was added to ∼20 mg of lyophilized cells for protein extraction. 25 µl of 5× sample buffer was then added, boiled for 5 min, and centrifuged at 15,000 rpm for 10 min. 10 µl of supernatant was loaded in duplicates for SDS-PAGE. Monoclonal antibodies against GFP (Roche) and tubulin (TAT1; Woods et al., 1989) were used.
Protein purification, mass spectrometry, gel filtration, and sedimentation velocity analytical ultracentrifugation
We used the previously described protocol for S-tag purification to identify the Rng8 binding partner (Liu et al., 2010). Approximately 1.5 g of lyophilized cells were broken by grinding with a mortar and pestle at room temperature until the cells became a homogenous fine powder. For protein extraction, the cell powder was thoroughly mixed with 20 ml of cold HK extraction buffer (25 mM Tris, pH 7.5, 1% NP-40, 300 mM NaCl, 5 mM EDTA, 15 mM EGTA, 60 mM β-Glycerophosphate, 500 µM Na3VO4, 10 mM NaF, 1 mM PMSF, 1 mM DTT, and protease inhibitors [Roche]). The cell extract was cleared by two rounds of centrifugation at 4°C (21,000 rpm for 30 min, 21,000 rpm for 10 min). The protein concentration was 10–15 mg/ml as measured using the Bradford method. Then cell extract was incubated with 300 mg S protein–conjugated agarose beads at 4°C for 2 h. The beads were collected by centrifugation at 4,000 rpm and washed once with an equal volume of HK extraction buffer, four times with an equal amount of washing buffer (25 mM Tris, pH 7.5, 300 mM NaCl, 5 mM EDTA, 500 µM Na3VO4, 10 mM NaF, 1 mM PMSF, and 1 mM DTT), and twice with 1 ml of washing buffer. To identify Rng8 binding partners, the proteins on the beads were released by boiling in 50 µl of 4× sample buffer for 5 min. For mass spectrometry analysis, the sample was run through the stacking SDS-PAGE gel and just into the resolving gel (Liu et al., 2010). All protein bands were excised as one sample and processed for mass spectrometry (Mass Spectrometry and Proteomics Facility, The Ohio State University).
To purify the Rng8–Rng9 complex from S. pombe, 3FLAG-Rng8 and Rng9-13Myc were simultaneously overexpressed by inducing the strain 3nmt1-3FLAG-rng8 3nmt1-rng9-13Myc myo51Δ in EMM5S medium for 48 h before collecting cells. The purification was similar to the purification of S-tagged Rng8 except for the following modifications: (1) ∼3 g of lyophilized cells were resuspended in 40 ml of HK extraction buffer; (2) cell extracts were incubated with 600 µl beads covalently coated with anti-FLAG antibody (catalogue No. F2426; Sigma-Aldrich) at 4°C for 1.5 h; and (3) to elute the proteins, beads were incubated with 1 ml 3FLAG peptide (catalogue No. F4799; Sigma-Aldrich) at a final concentration of 200 µg/ml in the washing buffer at 4°C for 30 min. Collected supernatant was concentrated to 300 µl for gel filtration using a protein concentrator (Vivaspin 500, catalogue no. VS0191; Sartorius Setim Biotech).
For estimating the size of the Rng8–Rng9 complex, a Superdex 200 column (28-9909-44; GE Healthcare) was first calibrated using the standard protein mixture (catalogue No. 151–1,901; Bio-Rad Laboratories). Then 300 µl of the purified Rng8–Rng9 complex was run alone or mixed with two of the purified standard proteins—γ-globulin (bovine) and myoglobin (horse)—and analyzed through the column using buffers containing 20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM DTT, 5% glycerol, and 0.01% NaN3 or 20 mM Tris, pH 7.5, 150 mM NaCl, and 1 mM TCEP for analytical ultracentrifugation.
Sedimentation velocity analytical ultracentrifugation experiments were performed at 20°C with an analytical ultracentrifuge (ProteomeLab XL-I; Beckman Coulter). Purified Rng8–Rng9 samples (in 20 mM Tris, 150 mM NaCl, and 1 mM TCEP, pH 7.5) were first clarified by centrifugation at 7,000 g for 8 min in a tabletop centrifuge, and ∼400 µl of each protein sample was loaded into two-channel centerpieces fitted with quartz windows in an eight-hole rotor. During centrifugation at 15,000 rpm at 20°C, samples were monitored by scanning absorbance at 280 nm and/or 230 nm versus radial location every 6–7 min. We used SEDFIT software (Schuck, 2000) to analyze the sedimentation profiles, and SEDNTERP software (Laue and Stafford, 1999) to calculate the density and viscosity of buffers from their compositions and the partial specific volume of the proteins from their amino acid composition.
Online supplemental material
Fig. S1 shows that Rng8 and Rng9 are CC proteins, and that rng8Δ, rng9Δ, and myo51Δ have similar cytokinesis defects. Fig. S2 shows that localizations of Myo52 and Rng8 are independent. Fig. S3 shows the synthetic interactions of myo51 mutations with myosin-II and ain1Δ mutations. Fig. S4 shows that Myo51 speckles cannot move along actin filaments. Fig. S5 shows that Rng8 and Rng9 interact to form a big protein complex. Table S1 lists the S. pombe strains used in this study. Table S2 lists the proteins identified in the mass spectrometry from the purification of Rng8-Stag. Video 1 shows Rng8 localization with a cell cycle marker SPB protein Sad1. Video 2 shows contractile ring assembly, maturation, and constriction in wt, rng8Δ, and myo51Δ cells expressing Rlc1-tdTomato at 36°C. Video 3 shows node condensation in wt and myo51Δ cells expressing Rlc1-tdTomato at 23°C. Video 4 shows contractile ring constriction and membrane invagination in wt, myo51Δ, myp2Δ, and myo51Δ myp2Δ cells. Video 5 shows contractile ring formation in wt and myo51Δ cells expressing Lifeact-mGFP. Video 6 shows Myo51-3GFP puncta moving along actin filament. Video 7 shows that Myo51-3YFP speckles do not move continuously and directionally in rng8Δ cells. Video 8 shows the nuclear movement in cells expressing Myo51N-GFP-Nup146. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201308146/DC1.
Supplementary Material
Acknowledgments
We thank Matthew Lord, Dimitrios Vavylonis, and Valarie Coffman for strains and communications on Myo51; Mohan Balasubramanian, Dannel McCollum, and Daniel Mulvihill for strains; Marina Bakhtina and Karin Musier-Forsyth for help with analytical ultracentrifugation; members of Stephen Osmani, Anita Hopper, James Hopper laboratories, and the Mass Spectrometry and Proteomics Facility at The Ohio State University for technical support; and I-Ju Lee for critical reading of the manuscript.
This work was supported by a Swiss National Science Foundation (SNF) research grant (31003A_138177) to S.G. Martin and the National Institutes of Health (NIH) grant GM086546 to J.-Q. Wu.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CC
- coiled-coil
- coIP
- coimmunoprecipitation
- DIC
- differential-interference contrast
- FL
- full length
- fps
- frames per second
- GTD
- globular tail domain
- IP
- immunoprecipitation
- Lat-A
- Latrunculin A
- MBC
- methyl benzimidazole-2-yl carbamate
- ROI
- region of interest
- SPB
- spindle pole body
- tdTomato
- tandem dimer Tomato
- wt
- wild type
References
- Bähler J., Steever A.B., Wheatley S., Wang Y., Pringle J.R., Gould K.L., McCollum D. 1998a. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J. Cell Biol. 143:1603–1616 10.1083/jcb.143.6.1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J., Wu J.-Q., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R. 1998b. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951 [DOI] [PubMed] [Google Scholar]
- Balasubramanian M.K., Bi E., Glotzer M. 2004. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr. Biol. 14:R806–R818 10.1016/j.cub.2004.09.022 [DOI] [PubMed] [Google Scholar]
- Barr F.A., Gruneberg U. 2007. Cytokinesis: placing and making the final cut. Cell. 131:847–860 10.1016/j.cell.2007.11.011 [DOI] [PubMed] [Google Scholar]
- Bezanilla M., Pollard T.D. 2000. Myosin-II tails confer unique functions in Schizosaccharomyces pombe: characterization of a novel myosin-II tail. Mol. Biol. Cell. 11:79–91 10.1091/mbc.11.1.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M., Forsburg S.L., Pollard T.D. 1997. Identification of a second myosin-II in Schizosaccharomyces pombe: Myp2p is conditionally required for cytokinesis. Mol. Biol. Cell. 8:2693–2705 10.1091/mbc.8.12.2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M., Wilson J.M., Pollard T.D. 2000. Fission yeast myosin-II isoforms assemble into contractile rings at distinct times during mitosis. Curr. Biol. 10:397–400 10.1016/S0960-9822(00)00420-6 [DOI] [PubMed] [Google Scholar]
- Bretscher A. 2003. Polarized growth and organelle segregation in yeast: the tracks, motors, and receptors. J. Cell Biol. 160:811–816 10.1083/jcb.200301035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheney R.E., O’Shea M.K., Heuser J.E., Coelho M.V., Wolenski J.S., Espreafico E.M., Forscher P., Larson R.E., Mooseker M.S. 1993. Brain myosin-V is a two-headed unconventional myosin with motor activity. Cell. 75:13–23 10.1016/0092-8674(93)90675-G [DOI] [PubMed] [Google Scholar]
- Chung S., Takizawa P.A. 2010. Multiple Myo4 motors enhance ASH1 mRNA transport in Saccharomyces cerevisiae. J. Cell Biol. 189:755–767 10.1083/jcb.200912011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman L.S., Okten Z., Rock R.S., Dawson J.F., Spudich J.A. 2005. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc. Natl. Acad. Sci. USA. 102:1419–1423 10.1073/pnas.0409487102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton J.E., Sammons M.R., Stark B.C., Hodges A.R., Lord M. 2010. Differential regulation of unconventional fission yeast myosins via the actin track. Curr. Biol. 20:1423–1431 10.1016/j.cub.2010.07.026 [DOI] [PubMed] [Google Scholar]
- Clayton J.E., Pollard L.W., Sckolnick M., Bookwalter C.S., Hodges A.R., Trybus K.M., Lord M. 2014. Fission yeast tropomyosin specifies directed transport of myosin-V along actin cables. Mol. Biol. Cell. 25:66–75 10.1091/mbc.E13-04-0200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman V.C., Wu J.-Q. 2012. Counting protein molecules using quantitative fluorescence microscopy. Trends Biochem. Sci. 37:499–506 10.1016/j.tibs.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman V.C., Nile A.H., Lee I-J., Liu H., Wu J.-Q. 2009. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol. Biol. Cell. 20:5195–5210 10.1091/mbc.E09-05-0428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman V.C., Wu P., Parthun M.R., Wu J.-Q. 2011. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J. Cell Biol. 195:563–572 10.1083/jcb.201106078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman V.C., Sees J.A., Kovar D.R., Wu J.-Q. 2013. The formins Cdc12 and For3 cooperate during contractile ring assembly in cytokinesis. J. Cell Biol. 203:101–114 10.1083/jcb.201305022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga R.R., Yonetani A., Chang F. 2006. Asymmetric microtubule pushing forces in nuclear centering. Curr. Biol. 16:1544–1550 10.1016/j.cub.2006.06.026 [DOI] [PubMed] [Google Scholar]
- Donovan K.W., Bretscher A. 2012. Myosin-V is activated by binding secretory cargo and released in coordination with Rab/exocyst function. Dev. Cell. 23:769–781 10.1016/j.devcel.2012.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle A., Martín-García R., Coulton A.T., Bagley S., Mulvihill D.P. 2009. Fission yeast Myo51 is a meiotic spindle pole body component with discrete roles during cell fusion and spore formation. J. Cell Sci. 122:4330–4340 10.1242/jcs.055202 [DOI] [PubMed] [Google Scholar]
- Dunn B.D., Sakamoto T., Hong M.S., Sellers J.R., Takizawa P.A. 2007. Myo4p is a monomeric myosin with motility uniquely adapted to transport mRNA. J. Cell Biol. 178:1193–1206 10.1083/jcb.200707080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eves P.T., Jin Y., Brunner M., Weisman L.S. 2012. Overlap of cargo binding sites on myosin V coordinates the inheritance of diverse cargoes. J. Cell Biol. 198:69–85 10.1083/jcb.201201024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Luo J., Nishihama R., Wloka C., Dravis C., Travaglia M., Iwase M., Vallen E.A., Bi E. 2010. Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J. Cell Biol. 191:1333–1350 10.1083/jcb.201005134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachet Y., Tournier S., Millar J.B., Hyams J.S. 2004. Mechanism controlling perpendicular alignment of the spindle to the axis of cell division in fission yeast. EMBO J. 23:1289–1300 10.1038/sj.emboj.7600156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. 2005. The molecular requirements for cytokinesis. Science. 307:1735–1739 10.1126/science.1096896 [DOI] [PubMed] [Google Scholar]
- Golomb L., Abu-Abied M., Belausov E., Sadot E. 2008. Different subcellular localizations and functions of Arabidopsis myosin VIII. BMC Plant Biol. 8:3 10.1186/1471-2229-8-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould K.L., Simanis V. 1997. The control of septum formation in fission yeast. Genes Dev. 11:2939–2951 10.1101/gad.11.22.2939 [DOI] [PubMed] [Google Scholar]
- Grallert A., Martín-García R., Bagley S., Mulvihill D.P. 2007. In vivo movement of the type V myosin Myo52 requires dimerisation but is independent of the neck domain. J. Cell Sci. 120:4093–4098 10.1242/jcs.012468 [DOI] [PubMed] [Google Scholar]
- Gross S.P., Vershinin M., Shubeita G.T. 2007. Cargo transport: two motors are sometimes better than one. Curr. Biol. 17:R478–R486 10.1016/j.cub.2007.04.025 [DOI] [PubMed] [Google Scholar]
- Hales K.G., Bi E., Wu J.-Q., Adam J.C., Yu I-C., Pringle J.R. 1999. Cytokinesis: an emerging unified theory for eukaryotes? Curr. Opin. Cell Biol. 11:717–725 10.1016/S0955-0674(99)00042-3 [DOI] [PubMed] [Google Scholar]
- Hammer J.A., III, Sellers J.R. 2012. Walking to work: roles for class V myosins as cargo transporters. Nat. Rev. Mol. Cell Biol. 13:13–26 http://dx.doi.org/10.1038/nrm3248 [DOI] [PubMed] [Google Scholar]
- Heuck A., Du T.G., Jellbauer S., Richter K., Kruse C., Jaklin S., Müller M., Buchner J., Jansen R.P., Niessing D. 2007. Monomeric myosin V uses two binding regions for the assembly of stable translocation complexes. Proc. Natl. Acad. Sci. USA. 104:19778–19783 10.1073/pnas.0706780104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heym R.G., Zimmermann D., Edelmann F.T., Israel L., Ökten Z., Kovar D.R., Niessing D. 2013. In vitro reconstitution of an mRNA-transport complex reveals mechanisms of assembly and motor activation. J. Cell Biol. 203:971–984 10.1083/jcb.201302095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges A.R., Krementsova E.B., Trybus K.M. 2008. She3p binds to the rod of yeast myosin V and prevents it from dimerizing, forming a single-headed motor complex. J. Biol. Chem. 283:6906–6914 10.1074/jbc.M708865200 [DOI] [PubMed] [Google Scholar]
- Hodges A.R., Bookwalter C.S., Krementsova E.B., Trybus K.M. 2009. A nonprocessive class V myosin drives cargo processively when a kinesin- related protein is a passenger. Curr. Biol. 19:2121–2125 10.1016/j.cub.2009.10.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges A.R., Krementsova E.B., Bookwalter C.S., Fagnant P.M., Sladewski T.E., Trybus K.M. 2012. Tropomyosin is essential for processive movement of a class V myosin from budding yeast. Curr. Biol. 22:1410–1416 10.1016/j.cub.2012.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Huang Y., Yu H., Subramanian D., Padmanabhan A., Thadani R., Tao Y., Tang X., Wedlich-Soldner R., Balasubramanian M.K. 2012. Nonmedially assembled F-actin cables incorporate into the actomyosin ring in fission yeast. J. Cell Biol. 199:831–847 10.1083/jcb.201209044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krementsova E.B., Hodges A.R., Bookwalter C.S., Sladewski T.E., Travaglia M., Sweeney H.L., Trybus K.M. 2011. Two single-headed myosin V motors bound to a tetrameric adapter protein form a processive complex. J. Cell Biol. 195:631–641 10.1083/jcb.201106146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krendel M., Mooseker M.S. 2005. Myosins: tails (and heads) of functional diversity. Physiology (Bethesda). 20:239–251 10.1152/physiol.00014.2005 [DOI] [PubMed] [Google Scholar]
- Kruse C., Jaedicke A., Beaudouin J., Bohl F., Ferring D., Guttler T., Ellenberg J., Jansen R.P. 2002. Ribonucleoprotein-dependent localization of the yeast class V myosin Myo4p. J. Cell Biol. 159:971–982 10.1083/jcb.200207101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D., Zhao R., Wu J.-Q. 2010. Mechanisms of contractile-ring assembly in fission yeast and beyond. Semin. Cell Dev. Biol. 21:892–898 10.1016/j.semcdb.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D., Coffman V.C., Lee I-J., Wu J.-Q. 2011. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J. Cell Biol. 192:1005–1021 10.1083/jcb.201008171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D., Ojkic N., Vavylonis D., Wu J.-Q. 2012. α-Actinin and fimbrin cooperate with myosin II to organize actomyosin bundles during contractile-ring assembly. Mol. Biol. Cell. 23:3094–3110 10.1091/mbc.E12-02-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue T.M., Stafford W.F., III 1999. Modern applications of analytical ultracentrifugation. Annu. Rev. Biophys. Biomol. Struct. 28:75–100 10.1146/annurev.biophys.28.1.75 [DOI] [PubMed] [Google Scholar]
- Lee I-J., Wu J.-Q. 2012. Characterization of Mid1 domains for targeting and scaffolding in fission yeast cytokinesis. J. Cell Sci. 125:2973–2985 10.1242/jcs.102574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I-J., Coffman V.C., Wu J.-Q. 2012. Contractile-ring assembly in fission yeast cytokinesis: Recent advances and new perspectives. Cytoskeleton (Hoboken). 69:751–763 10.1002/cm.21052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.-L., Osmani A.H., Ukil L., Son S., Markossian S., Shen K.F., Govindaraghavan M., Varadaraj A., Hashmi S.B., De Souza C.P., Osmani S.A. 2010. Single-step affinity purification for fungal proteomics. Eukaryot. Cell. 9:831–833 10.1128/EC.00032-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti L., Martin S.G. 2011. Shaping fission yeast cells by rerouting actin-based transport on microtubules. Curr. Biol. 21:2064–2069 10.1016/j.cub.2011.10.033 [DOI] [PubMed] [Google Scholar]
- Lo Presti L., Chang F., Martin S.G. 2012. Myosin Vs organize actin cables in fission yeast. Mol. Biol. Cell. 23:4579–4591 10.1091/mbc.E12-07-0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M., Laves E., Pollard T.D. 2005. Cytokinesis depends on the motor domains of myosin-II in fission yeast but not in budding yeast. Mol. Biol. Cell. 16:5346–5355 10.1091/mbc.E05-07-0601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A., Arai R., Yashiroda Y., Shirai A., Kamata A., Sekido S., Kobayashi Y., Hashimoto A., Hamamoto M., Hiraoka Y., et al. 2006. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 24:841–847 10.1038/nbt1222 [DOI] [PubMed] [Google Scholar]
- May K.M., Win T.Z., Hyams J.S. 1998. Yeast myosin II: a new subclass of unconventional conventional myosins? Cell Motil. Cytoskeleton. 39:195–200 [DOI] [PubMed] [Google Scholar]
- Mehta A.D., Rock R.S., Rief M., Spudich J.A., Mooseker M.S., Cheney R.E. 1999. Myosin-V is a processive actin-based motor. Nature. 400:590–593 10.1038/23072 [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Motegi F., Nakano K., Mabuchi I. 2000. Molecular mechanism of myosin-II assembly at the division site in Schizosaccharomyces pombe. J. Cell Sci. 113:1813–1825 [DOI] [PubMed] [Google Scholar]
- Motegi F., Mishra M., Balasubramanian M.K., Mabuchi I. 2004. Myosin-II reorganization during mitosis is controlled temporally by its dephosphorylation and spatially by Mid1 in fission yeast. J. Cell Biol. 165:685–695 10.1083/jcb.200402097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Richter K., Heuck A., Kremmer E., Buchner J., Jansen R.P., Niessing D. 2009. Formation of She2p tetramers is required for mRNA binding, mRNP assembly, and localization. RNA. 15:2002–2012 10.1261/rna.1753309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvihill D.P., Pollard P.J., Win T.Z., Hyams J.S. 2001. Myosin V-mediated vacuole distribution and fusion in fission yeast. Curr. Biol. 11:1124–1127 10.1016/S0960-9822(01)00322-0 [DOI] [PubMed] [Google Scholar]
- Mulvihill D.P., Edwards S.R., Hyams J.S. 2006. A critical role for the type V myosin, Myo52, in septum deposition and cell fission during cytokinesis in Schizosaccharomyces pombe. Cell Motil. Cytoskeleton. 63:149–161 10.1002/cm.20113 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Nakamura-Kubo M., Hirata A., Shimoda C. 2001. The Schizosaccharomyces pombe spo3+ gene is required for assembly of the forespore membrane and genetically interacts with psy1(+)-encoding syntaxin-like protein. Mol. Biol. Cell. 12:3955–3972 10.1091/mbc.12.12.3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B.J., Tomasevic N., Russell A., Pierce D.W., Jia Z., McCormick C.D., Hartman J., Sakowicz R., Pollard T.D. 2009. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 460:1031–1034 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odronitz F., Kollmar M. 2007. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 8:R196 10.1186/gb-2007-8-9-r196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti A., Chang F. 2000. Analysis of mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission yeast. Mol. Biol. Cell. 11:2757–2773 10.1091/mbc.11.8.2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkova N., Jin Y., Ramaswamy S., Weisman L.S. 2006. Structural basis for myosin V discrimination between distinct cargoes. EMBO J. 25:693–700 10.1038/sj.emboj.7600965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Wu J.-Q. 2010. Understanding cytokinesis: lessons from fission yeast. Nat. Rev. Mol. Cell Biol. 11:149–155 10.1038/nrm2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D., Legesse-Miller A., Gao L., Dong Y., Bretscher A. 2004. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 20:559–591 10.1146/annurev.cellbio.20.010403.103108 [DOI] [PubMed] [Google Scholar]
- Reck-Peterson S.L., Tyska M.J., Novick P.J., Mooseker M.S. 2001. The yeast class V myosins, Myo2p and Myo4p, are nonprocessive actin-based motors. J. Cell Biol. 153:1121–1126 10.1083/jcb.153.5.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rief M., Rock R.S., Mehta A.D., Mooseker M.S., Cheney R.E., Spudich J.A. 2000. Myosin-V stepping kinetics: a molecular model for processivity. Proc. Natl. Acad. Sci. USA. 97:9482–9486 10.1073/pnas.97.17.9482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith R.H., Gould K.L. 2008. Stepping into the ring: the SIN takes on contractile ring assembly. Genes Dev. 22:3082–3088 10.1101/gad.1748908 [DOI] [PubMed] [Google Scholar]
- Rout M.P., Aitchison J.D., Suprapto A., Hjertaas K., Zhao Y., Chait B.T. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635–652 10.1083/jcb.148.4.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf R., Bittins C.M., Gerdes H.H. 2011. The role of myosin V in exocytosis and synaptic plasticity. J. Neurochem. 116:177–191 10.1111/j.1471-4159.2010.07110.x [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Amitani I., Yokota E., Ando T. 2000. Direct observation of processive movement by individual myosin V molecules. Biochem. Biophys. Res. Commun. 272:586–590 10.1006/bbrc.2000.2819 [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Wang F., Schmitz S., Xu Y., Xu Q., Molloy J.E., Veigel C., Sellers J.R. 2003. Neck length and processivity of myosin V. J. Biol. Chem. 278:29201–29207 10.1074/jbc.M303662200 [DOI] [PubMed] [Google Scholar]
- Šamaj J., Read N.D., Volkmann D., Menzel D., Baluska F. 2005. The endocytic network in plants. Trends Cell Biol. 15:425–433 10.1016/j.tcb.2005.06.006 [DOI] [PubMed] [Google Scholar]
- Samejima I., Miller V.J., Rincon S.A., Sawin K.E. 2010. Fission yeast Mto1 regulates diversity of cytoplasmic microtubule organizing centers. Curr. Biol. 20:1959–1965 10.1016/j.cub.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Bowers B., Varma A., Roh D.H., Cabib E. 2002. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J. Cell Sci. 115:293–302 [DOI] [PubMed] [Google Scholar]
- Schuck P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78:1606–1619 10.1016/S0006-3495(00)76713-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladewski T.E., Bookwalter C.S., Hong M.S., Trybus K.M. 2013. Single-molecule reconstitution of mRNA transport by a class V myosin. Nat. Struct. Mol. Biol. 20:952–957 10.1038/nsmb.2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark B.C., Sladewski T.E., Pollard L.W., Lord M. 2010. Tropomyosin and myosin-II cellular levels promote actomyosin ring assembly in fission yeast. Mol. Biol. Cell. 21:989–1000 10.1091/mbc.E09-10-0852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P., Kunze R., Flemming D., Höpfner D., Diepholz M., Philippsen P., Böttcher B., Hurt E. 2007. Molecular basis for the functional interaction of dynein light chain with the nuclear-pore complex. Nat. Cell Biol. 9:788–796 10.1038/ncb1604 [DOI] [PubMed] [Google Scholar]
- Tebbs I.R., Pollard T.D. 2013. Separate roles of IQGAP Rng2p in forming and constricting the Schizosaccharomyces pombe cytokinetic contractile ring. Mol. Biol. Cell. 24:1904–1917 10.1091/mbc.E12-10-0775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolić-Nørrelykke I.M., Sacconi L., Stringari C., Raabe I., Pavone F.S. 2005. Nuclear and division-plane positioning revealed by optical micromanipulation. Curr. Biol. 15:1212–1216 10.1016/j.cub.2005.05.052 [DOI] [PubMed] [Google Scholar]
- Tominaga M., Nakano A. 2012. Plant-specific myosin XI, a molecular perspective. Front. Plant Sci. 3:211 10.3389/fpls.2012.00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth J., Kovács M., Wang F., Nyitray L., Sellers J.R. 2005. Myosin V from Drosophila reveals diversity of motor mechanisms within the myosin V family. J. Biol. Chem. 280:30594–30603 10.1074/jbc.M505209200 [DOI] [PubMed] [Google Scholar]
- Tran P.T., Marsh L., Doye V., Inoué S., Chang F. 2001. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 153:397–412 10.1083/jcb.153.2.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D., Wu J.-Q., Hao S., O’Shaughnessy B., Pollard T.D. 2008. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 319:97–100 10.1126/science.1151086 [DOI] [PubMed] [Google Scholar]
- Wagner W., Bielli P., Wacha S., Ragnini-Wilson A. 2002. Mlc1p promotes septum closure during cytokinesis via the IQ motifs of the vesicle motor Myo2p. EMBO J. 21:6397–6408 10.1093/emboj/cdf650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M.L., Burgess S.A., Sellers J.R., Wang F., Hammer J.A., III, Trinick J., Knight P.J. 2000. Two-headed binding of a processive myosin to F-actin. Nature. 405:804–807 10.1038/35015592 [DOI] [PubMed] [Google Scholar]
- Warshaw D.M., Kennedy G.G., Work S.S., Krementsova E.B., Beck S., Trybus K.M. 2005. Differential labeling of myosin V heads with quantum dots allows direct visualization of hand-over-hand processivity. Biophys. J. 88:L30–L32 10.1529/biophysj.105.061903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Watanabe T.M., Sato O., Awata J., Homma K., Umeki N., Higuchi H., Ikebe R., Ikebe M. 2008. Human myosin Vc is a low duty ratio nonprocessive motor. J. Biol. Chem. 283:10581–10592 10.1074/jbc.M707657200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win T.Z., Gachet Y., Mulvihill D.P., May K.M., Hyams J.S. 2001. Two type V myosins with non-overlapping functions in the fission yeast Schizosaccharomyces pombe: Myo52 is concerned with growth polarity and cytokinesis, Myo51 is a component of the cytokinetic actin ring. J. Cell Sci. 114:69–79 [DOI] [PubMed] [Google Scholar]
- Wloka C., Vallen E.A., Thé L., Fang X., Oh Y., Bi E. 2013. Immobile myosin-II plays a scaffolding role during cytokinesis in budding yeast. J. Cell Biol. 200:271–286 10.1083/jcb.201208030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe B.A., Gould K.L. 2005. Split decisions: coordinating cytokinesis in yeast. Trends Cell Biol. 15:10–18 10.1016/j.tcb.2004.11.006 [DOI] [PubMed] [Google Scholar]
- Woods A., Sherwin T., Sasse R., MacRae T.H., Baines A.J., Gull K. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93:491–500 [DOI] [PubMed] [Google Scholar]
- Wu J.-Q., Pollard T.D. 2005. Counting cytokinesis proteins globally and locally in fission yeast. Science. 310:310–314 10.1126/science.1113230 [DOI] [PubMed] [Google Scholar]
- Wu J.-Q., Bähler J., Pringle J.R. 2001. Roles of a fimbrin and an α-actinin-like protein in fission yeast cell polarization and cytokinesis. Mol. Biol. Cell. 12:1061–1077 10.1091/mbc.12.4.1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-Q., Kuhn J.R., Kovar D.R., Pollard T.D. 2003. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell. 5:723–734 10.1016/S1534-5807(03)00324-1 [DOI] [PubMed] [Google Scholar]
- Wu J.-Q., Sirotkin V., Kovar D.R., Lord M., Beltzner C.C., Kuhn J.R., Pollard T.D. 2006. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J. Cell Biol. 174:391–402 10.1083/jcb.200602032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-Q., McCormick C.D., Pollard T.D. 2008. Chapter 9: Counting proteins in living cells by quantitative fluorescence microscopy with internal standards. Methods Cell Biol. 89:253–273 10.1016/S0091-679X(08)00609-2 [DOI] [PubMed] [Google Scholar]
- Wu P., Zhao R., Ye Y., Wu J.-Q. 2011. Roles of the DYRK kinase Pom2 in cytokinesis, mitochondrial morphology, and sporulation in fission yeast. PLoS ONE. 6:e28000 10.1371/journal.pone.0028000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Lee I-J., Runge K.W., Wu J.-Q. 2012. Roles of putative Rho-GEF Gef2 in division-site positioning and contractile-ring function in fission yeast cytokinesis. Mol. Biol. Cell. 23:1181–1195 10.1091/mbc.E11-09-0800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A., Forkey J.N., McKinney S.A., Ha T., Goldman Y.E., Selvin P.R. 2003. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 300:2061–2065 10.1126/science.1084398 [DOI] [PubMed] [Google Scholar]
- Yin H., Pruyne D., Huffaker T.C., Bretscher A. 2000. Myosin V orientates the mitotic spindle in yeast. Nature. 406:1013–1015 10.1038/35023024 [DOI] [PubMed] [Google Scholar]
- Yu J.H., Crevenna A.H., Bettenbühl M., Freisinger T., Wedlich-Söldner R. 2011. Cortical actin dynamics driven by formins and myosin V. J. Cell Sci. 124:1533–1541 10.1242/jcs.079038 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.