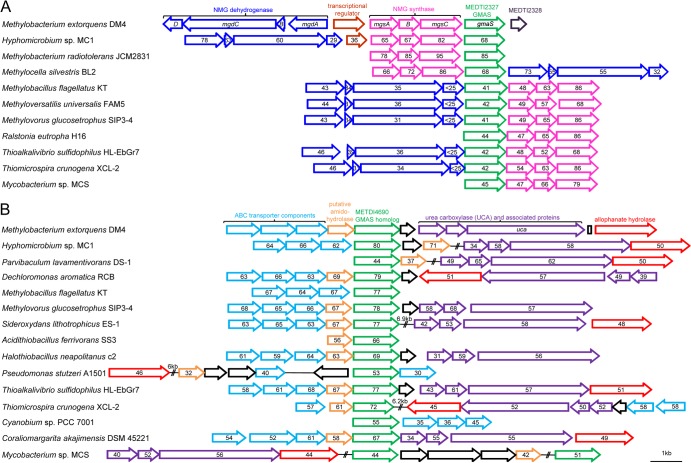
FIG 5.
Gene organization and conservation of gmaS homolog-containing genomic regions for sequences closely related to METDI2327 (A) and METI4690 (B), respectively. Gene orientation and linkage are indicated with arrows, and double slashes indicate a distance of at least 10 kb between the genome regions considered. UCA-associated proteins 1 and 2 are encoded by a tandem of homologous, uncharacterized genes with conserved domains TIGR03424 and TIGR03425, respectively. Alphaproteobacteria class members (complete genome accession numbers are in parentheses): M. extorquens DM4 (NC_012988), Hyphomicrobium sp. strain MC1 (NC_015717), Methylobacterium radiotolerans JCM2831 (NC_010505), M. silvestris BL2 (NC_011666), and Parvibaculum lavamentivorans DS-1 (NC_009719). Betaproteobacteria class members: Dechloromonas aromatica RCB (NC_007298), Methylobacillus flagellatus KT (NC_007947), M. universalis FAM5 (NZ_AFHG00000000), Methylovorus glucosetrophus SIP3-4 (NC_012969), Sideroxydans lithotrophicus ES-1 (NC_013959), and Ralstonia eutropha H16 (NC_008314). Gammaproteobacteria class members: Acidithiobacillus ferrivorans SS3 (NC_015942), Halothiobacillus neapolitanus c2 (NC_013422), Pseudomonas stutzeri A1501 (NC_009434), Thioalkalivibrio sulfidophilus HL-EbGr7 (NC_011901), and Thiomicrospira crunogena XCL-2 (NC_007520). Cyanobacteria class member: Cyanobium sp. strain PCC 7001 (NZ_DS990556). Verrucomicrobia class member: Coraliomargarita akajimensis DSM 45221 (NC_014008). Actinobacteria class member: Mycobacterium sp. strain MCS (NC_008147).