Figure 2. cGMP binding pocket of CNB-B and its comparison with the PKA:cAMP complex.
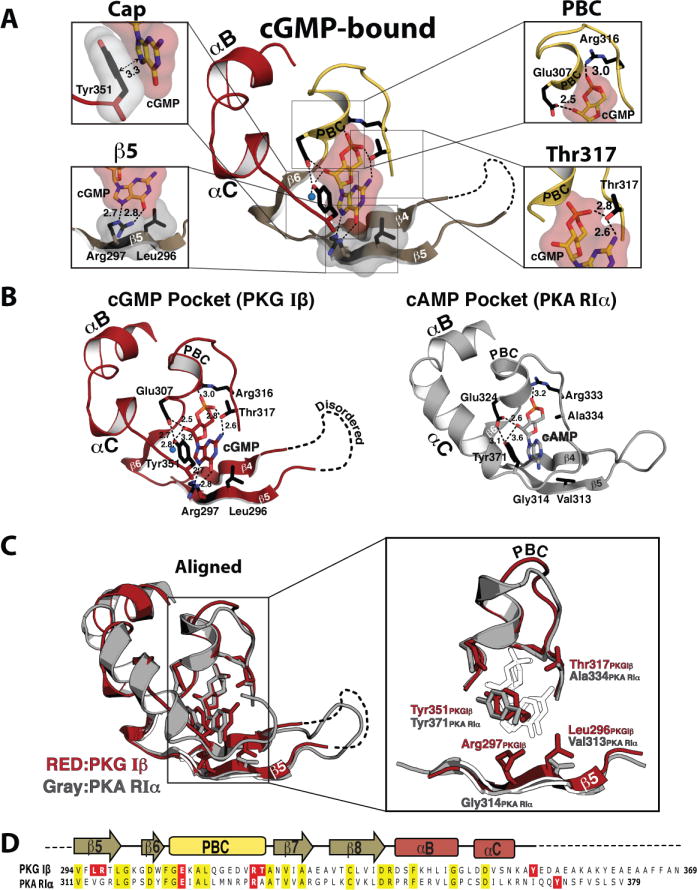
(A) Detailed interactions between cGMP and CNB-B. Zoomed in views for each cGMP binding site are shown on either side. The individual cGMP interacting residues are shown, with the following color theme: side chain carbon, black; oxygen, red; nitrogen, blue. The cyclic nucleotide, β5 and capping residues are shown with transparent surface. Contacts are shown as dotted lines with their distances (Å) (B) Comparison between the PKG Iβ cGMP binding pocket and PKA RIα cAMP pocket. The cGMP pocket of PKG Iβ in red is shown on the left panel and the cAMP binding pocket of PKA RIα (PDB code: 1RGS) in gray is shown on the right. Key cyclic nucleotide binding residues are shown as sticks. An ordered water molecule is shown as blue sphere. (C) Superposition of two structures shown in B are shown on the left, with a zoomed in view on the right (cyclic nucleotides are showm as white sticks). (D) Sequence Alignments of CNB-B from PKG Iβ and PKA RIα. Conserved residues are shaded in yellow and cyclic nucleotide interacting residues through their side chains are shaded in red for both proteins.
