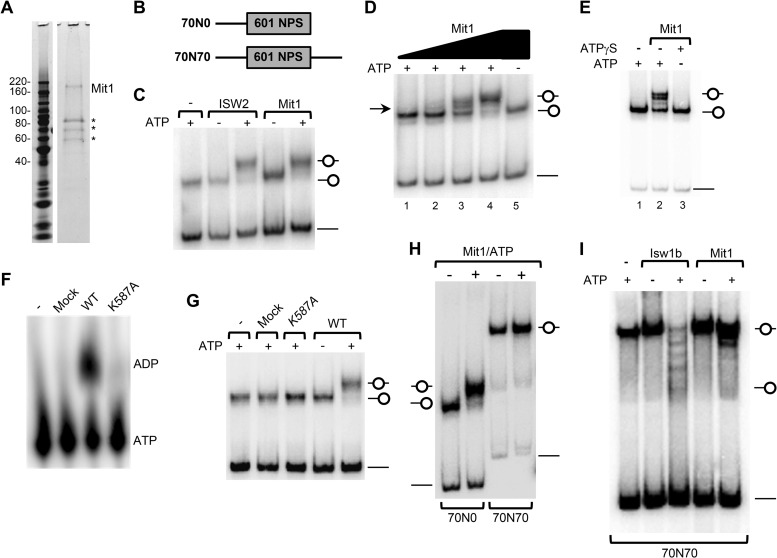
FIG 3.
Mit1 is a directional ATP-dependent nucleosome remodeling factor. (A) Purification of Mit1. An aliquot of the final purification of 3×FLAG-Mit1–3×V5 expressed in mit1Δ cells was analyzed by SDS-PAGE and SYPRO ruby staining. Mit1 was identified as the approximately 170-kDa band with 49% sequence coverage by mass spectrometry. Asterisks represent Mit1 breakdown products and heat shock proteins. See Table S4 in the supplemental material for more details. (B) Schematic representation of short mononucleosomes reconstituted for remodeling experiments. Nucleosomes were positioned by the Widom 601 nucleosome positioning sequence. (C) Mit1 can remodel a mononucleosomal substrate. End-positioned (70N0) mononucleosomes (30 nM) were incubated with ScISW2 (5.0 nM) or S. pombe Mit1 (SpMit1) (2.5 nM) in the presence or absence of ATP and resolved on a native 5% polyacrylamide gel. (D) Titration of Mit1 protein reveals intermediate remodeling events. Nucleosomes were remodeled as for panel C with a titration of Mit1 (0.3 nM, 0.6 nM, 1.25 nM, and 2.5 nM). (E) Mit1 utilizes ATP hydrolysis to remodel a mononucleosomal substrate. Nucleosomes were remodeled as for panel C on addition of ATP but not on addition of the nonhydrolyzable analogue, ATPγS. (F) Mit1 but not Mit1K587A can hydrolyze ATP. ATP hydrolysis assay comparing the activity of wild-type Mit1 to that of Mit1K587A, a mock purification, and a buffer-only control. (G) Mit1K587A cannot mobilize nucleosomes. Nucleosome mobilization assays as performed for panel C, comparing octamer mobilization by wild-type Mit1, Mit1K587A, and mock purifications. (H) Mit1 is a directional remodeler. Nucleosome mobilization assay comparing the remodeling of end-positioned (70N0) and center-positioned (70N70) nucleosomes incubated with or without Mit1 (2.5 nM) and ATP. (I) Comparison of Mit1 and Isw1b remodeling. Center-positioned mononucleosomes (70N70; 30 nM) were incubated with purified ScIsw1b (5 nM) or Mit1 (5 nM) in the presence or absence of ATP.