FIG 4.
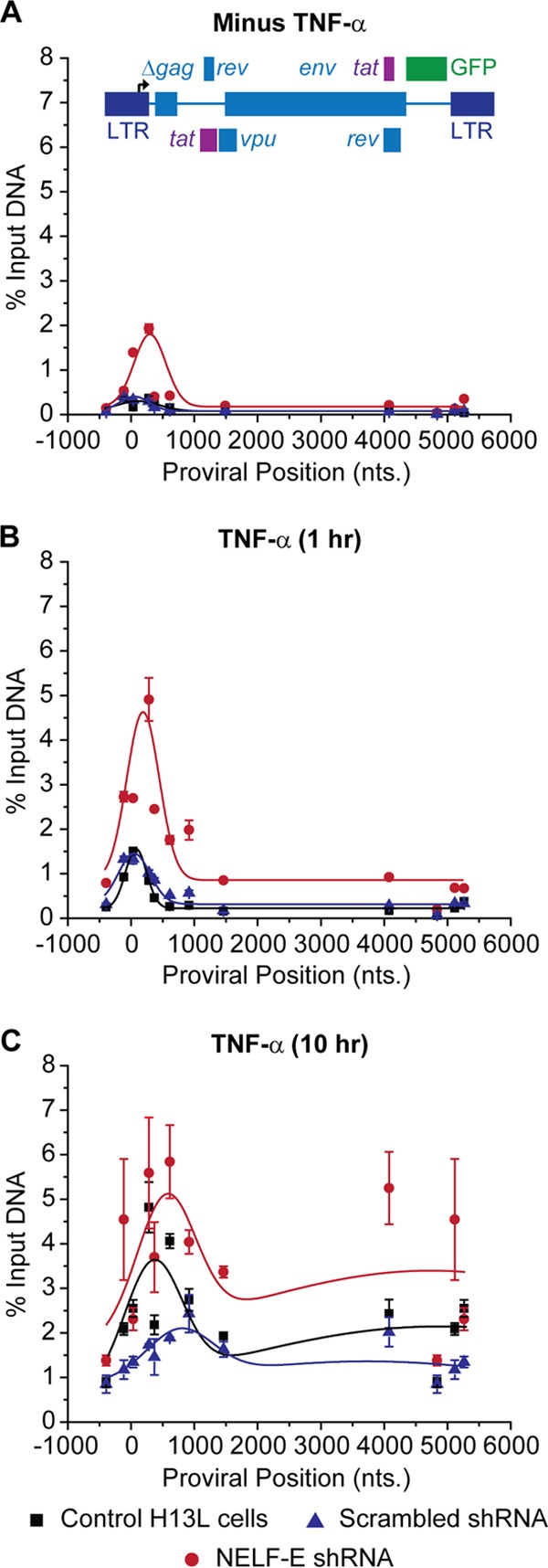
Distribution of RNAP II on the HIV genome following knockdown of NELF-E. ChIP analyses were performed using NELF-E shRNA (clone N2G10) and its scrambled control (clone N2F2) in unstimulated cells (minus TNF-α) (A), cells stimulated with 2 ng/ml TNF-α for 1 h (B), and cells stimulated with 2 ng/ml TNF-α for 10 h (C). Error bars represent the standard deviations of triplicate real-time PCR determinations for each primer set.
