FIG 5.
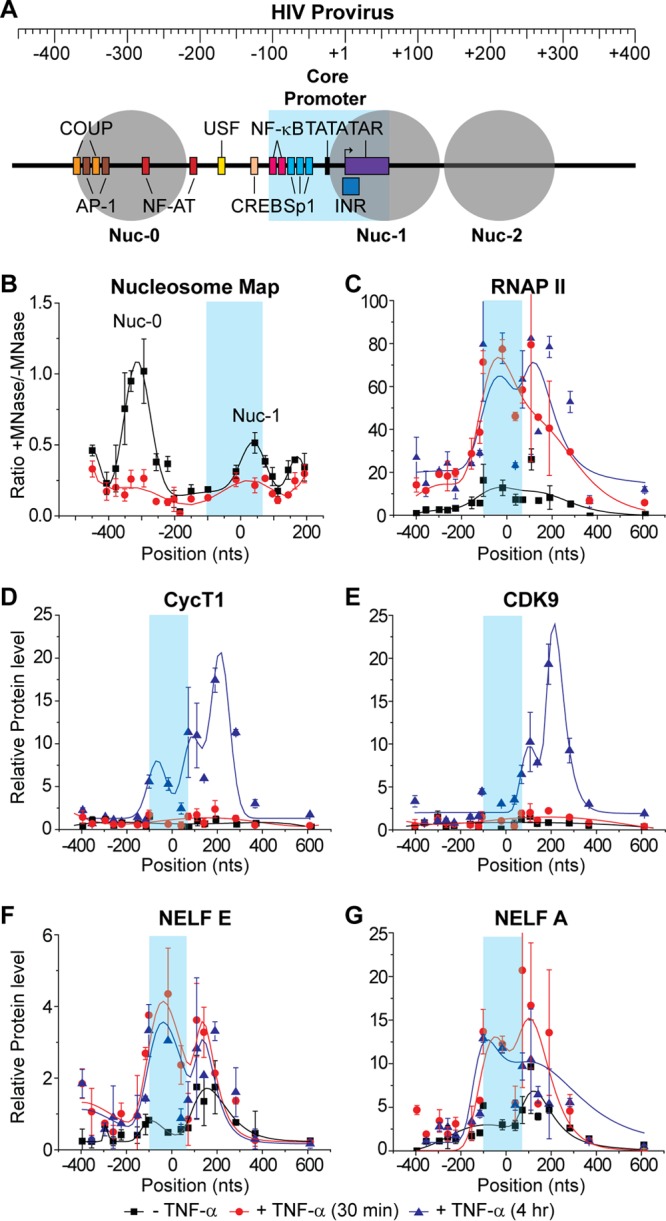
Distribution of RNAP II and associated transcription factors on the HIV genome following TNF-α activation. ChIP assays were used to measure the density of RNA polymerase along the proviral genome from the core promoter region in the LTR (shaded in blue) to 600 nt downstream of the transcription start site. (A) Schematic map of the HIV LTR showing nucleosomes and transcription factor binding sites. (B) Micrococcal nuclease protection assay. (C to G) ChIP analyses were performed using antibodies against the indicated proteins: RNAP II, CycT1, CDK9, NELF-E, and NELF-A. Error bars represent standard deviations of triplicate real-time PCR determinations for each primer set.
