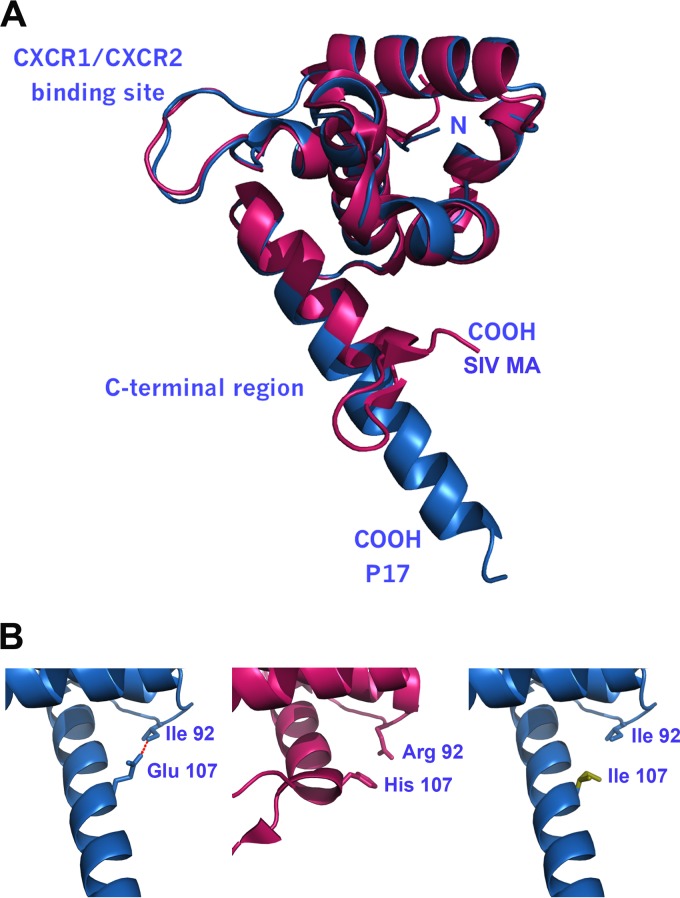
FIG 7.
Similarities and differences in the C-terminal regions of p17, SIV MA, and S75X. (A) Three-dimensional alignment of p17 (blue) and SIV MA (red). Both proteins have a nearly identical three-dimensional structure in the N-terminal region down to residue 107. The conformation of the AT20 loops of p17 and SIV MA is preserved for binding to CXCR1 and CXCR2. (B) The α-helix H5 in p17 is bound to the loop following H4 by an H-bond of Ile92 to Glu107 (red), thus stabilizing the conformation between H5 and the rest of the protein (left panel). In the C-terminal end of SIV MA the residue at position 107 is His, and residues 110 to 117 build a β-sheet (middle panel). The model shows that there is no way to keep H5 in place when Glu107 is mutated to Ile, as in S75X (right panel).