ABSTRACT
Interferons (IFNs) are cytokines produced by host cells in response to the infection with pathogens. By binding to the corresponding receptors, IFNs trigger different pathways to block intracellular replication and growth of pathogens and to impede the infection of surrounding cells. Due to their key role in host defense against viral infections, as well as for clinical therapies, the IFN responses and regulation mechanisms are well studied. However, studies of type I IFNs have mainly focused on alpha interferon (IFN-α) and IFN-β subtypes. Knowledge of IFN-κ and IFN-ω is limited. Moreover, most studies are performed in humans or mouse models but not in the original host of zoonotic pathogens. Bats are important reservoirs and transmitters of zoonotic viruses such as lyssaviruses. A few studies have shown an antiviral activity of IFNs in fruit bats. However, the function of type I IFNs against lyssaviruses in bats has not been studied yet. Here, IFN-κ and IFN-ω genes from the European serotine bat, Eptesicus serotinus, were cloned and functionally characterized. E. serotinus IFN-κ and IFN-ω genes are intronless and well conserved between microchiropteran species. The promoter regions of both genes contain essential regulatory elements for transcription factors. In vitro studies indicated a strong activation of IFN signaling by recombinant IFN-ω, whereas IFN-κ displayed weaker activation. Noticeably, both IFNs inhibit to different extents the replication of different lyssaviruses in susceptible bat cell lines. The present study provides functional data on the innate host defense against lyssaviruses in endangered European bats.
IMPORTANCE We describe here for the first time the molecular and functional characterization of two type I interferons (IFN-κ and -ω) from European serotine bat (Eptesicus serotinus). The importance of this study is mainly based on the fact that very limited information about the early innate immune response against bat lyssaviruses in their natural host serotine bats is yet available. Generally, whereas the antiviral activity of other type I interferons is well studied, the functional involvement of IFN-κ and -ω has not yet been investigated.
INTRODUCTION
Worldwide, more than 1,200 different species of bats (Chiroptera) have been described; thus, bats represent the second most diverse mammalian order (1). Bats have been identified as reservoirs for a plethora of viruses (2). Some, such as severe acute respiratory syndrome coronavirus (SARS-CoV) and Hendra virus, are associated with recent human and animal epidemics. Thus, bats present a potential threat to public health (2–4). Bats of the genus Eptesicus can transmit lyssaviruses (5). Surprisingly, reports about epidemics inside bat populations are very limited, indicating that viral pathogens do not seem to induce major disease outbreaks and that certain pathogens, such as rabies virus (RABV), are better controlled within bats (2, 6, 7). Therefore, it is uncertain whether a coevolution of lyssaviruses and the bat immune system resulted in a partial resistance against infections due to the specific adaptation of the innate immune system of bats (8–11).
Interferons (IFNs) are the first-line defenders against lyssavirus infections (12–15). IFNs are produced by host cells in response to pathogens such as viruses, bacteria, fungi, and parasites and trigger protective immune mechanisms. Vertebrate IFNs are classified into three classes: type I IFNs, type II IFNs, and type III IFNs. Type I IFNs are the largest group in the IFN family. Due to their versatility, they have expanded and diverged into many distinct subfamilies: IFN-α, IFN-β, IFN-κ, IFN-ω, IFN-ε, IFN-δ, IFN-ζ, and IFN-τ, as well as a potentially new IFN, “IFNX” (16). With a conserved intronless structure and colocalized loci in the chromosome, type I IFNs are considered to have arisen and expanded from gene duplications (17, 18). Studies of their antiviral defenses and regulation mechanisms are rapidly mounting. Most of these studies focus on the IFN-α and IFN-β subtypes, and little is known about other subtypes, even in humans and in mouse models. IFNs seem to play a crucial role in host-virus interactions in bats (19–21). However, such studies have been limited to type I IFN-α and IFN-β from the Egyptian fruit bat (Rousettusae gyptiacus) and type II IFN-γ, type III IFN-λ1, and type III IFN-λ2 from the Australian black flying fox, Pteropus alecto (22–24). In addition to sequence characterization, IFN induction, the signaling pathway, and antiviral activity have all been investigated in cell lines derived from Eidolon helvum and P. alecto after viral infections (20–23). Similar to mammalian IFNs, P. alecto IFN-γ and IFN-λ2 showed antiviral activities against Semliki Forest virus and Hendra virus and against Pulau virus, respectively (22, 23).
Surprisingly, although bats are known as the principal vectors for RABV and other lyssaviruses, information on IFNs during lyssavirus infections is scarce (2). In Europe, five different bat species have been identified as potential reservoir hosts for lyssaviruses: E. serotinus, E. isabellinus (European bat lyssavirus type 1 [EBLV-1]), Myotis daubentonii, M. dasycneme (EBLV-2), M. nattereri (Bokeloh bat lyssavirus), and Miniopterus schreibersi (West Caucasian bat lyssavirus and Lleida bat lyssavirus) (25–28). Since E. serotinus, a the natural host for EBLV-1, is responsible for the vast majority of bat rabies cases reported in Europe (29), we focused on a functional characterization of IFN from this species.
Since we could clone first IFN-κ and IFN-ω genes from E. serotinus, we focused in the present study on the molecular and functional characterization of these two type I IFNs. After sequence analysis, recombinant IFN-κ and IFN-ω proteins were used in functional assays to investigate the initiation of the IFN signaling pathway and antilyssaviral activity against EBLV-1, EBLV-2, and RABV infections in E. serotinus-derived cell lines. Our analysis sheds light on the function of IFNs within cells of the natural host of RABV and other lyssaviruses and the virus-host interaction in these specific mammalian hosts.
MATERIALS AND METHODS
Animal sampling.
E. serotinus bats submitted for rabies diagnosis in the frame of enhanced passive surveillance (J. Schatz, unpublished data) were screened for their suitability to obtain immune system-related organ tissues. The spleens from two dead E. serotinus bats from the city of Berlin, Germany, that did not show any evidence of accelerated autolysis were removed and stored in RNAlater until further analysis.
Nucleic acid extraction from tissue.
Total RNA extraction of the E. serotinus spleen was performed using an RNeasy minikit (Qiagen, Germany) according to the manufacturer's protocol. Genomic DNA was also isolated from spleen tissue by using a DNeasy Blood & Tissue kit (Qiagen). The concentration and purity of RNA and genomic DNA were determined by using NanoDrop (Thermo, USA) and then stored at −80°C for further use.
PCR amplification of IFN-κ and IFN-ω genes from E. serotinus (esIFN-κ and esIFN-ω).
Since no appropriate genomic sequences for E. serotinus available were available, the following approach was used. IFN-κ from P. vampyrus (GenBank accession no. HM636500) and IFN-ω from Equus caballus (GenBank accession no. NM_001114536) were used as query sequences to search for the IFN-κ and IFN-ω genes, respectively, from a small brown bat, M. lucifugus, in the whole-genome sequence in the Ensembl database using the BLAST algorithm (http://www.ensembl.org/index.html). The exons from the matching scaffolds were then subjected to the National Center for Biotechnology Information (NCBI) BLAST programs blastN and blastX to confirm the identities of IFN-κ and IFN-ω.
Reverse transcription-PCR (RT-PCR) was performed to amplify the IFN-κ and IFN-ω cDNA fragments from E. serotinus RNA using primer pairs based on the sequences of IFN-κ and IFN-ω genes from M. lucifugus (Table 1). The reactions were prepared according to the manufacturer's instructions with a One-Step RT-PCR kit (Qiagen). PCR was performed using genomic DNA as the template with GoTaq Flexi DNA polymerase (Promega, USA) to obtain the DNA fragments. All of the PCR products were cloned into PCR2.1 vector (Invitrogen, USA) and transformed into Escherichia coli competent cells. Plasmids were extracted from positive clones and sequenced by using an Applied Biosystems 3130 genetic analyzer (Life Technologies, USA) at the Friedrich-Loeffler-Institut, Greifswald-Insel Riems, Germany.
TABLE 1.
Primers used in this study
| Primera | Sequence (5′–3′) |
|---|---|
| IFNκ-F | TGACCAATATGAGCAAATCA |
| IFNκ-R | AAACTGCTTATCCTGTGGAA |
| IFNω-F | GACCACGTCCAGCTCAGC |
| IFNω-R | GCGCAGTCACTGTATTTCTT |
| gwesIFNκ-F1 | CAAGGGGGCCTTCTATGAAATGTCCATG |
| gwesIFNκ-F2 | TTCACTCAACCCACCTTCCAACCCACTT |
| gwesIFNκ-R1 | CCCTGGTGTCTCCTCTCATCTCCTCCA |
| gwesIFNκ-R2 | AAGTGGTTGGAAGGTGGGTGGAGTGAA |
| gwesIFNκ-R3 | GAAGGCCCCCTTGATGTCTCTTTTCA |
| gwesIFNκ-R4 | AATATGCAGCAAGTCACAGCCCAGT |
| gwesIFNω-F1 | AGCACCATCTCCCCTCTCTTCTGTCTG |
| gwesIFNω-F2 | ATCTCTGACCTCCTGCCCACAGAGAAC |
| gwesIFNω-R1 | CAGGGCCATGAGTAGAGAGAGCAGGAG |
| gwesIFNω-R2 | CCTTGGGCCAGAAGTTCTCCTATGACC |
| gwesIFNω-R3 | GCCCTAATGGTTTGGCTCAGTGGAT |
| gwesIFNω-R4 | GGTGTGCAGGAGGCAGCTTATCAGT |
| AP1 | GTAATACGACTCACTATAGGGC |
| AP2 | ACTATAGGGCACGCGTGGT |
| PmalesIFNκ-F | AAGGATTTCAGAATTCCTGGGCTGTGACTTGCTG |
| PmalesIFNκ-R | TAGAGGATCCGAATTCTTATTTCCTTCTGAGTAGTTC |
| PmalesIFNω-F | AAGGATTTCAGAATTCTGTGACCTGCCTGAGGACC |
| PmalesIFNω-R | TAGAGGATCCGAATTCTCAAGGTGACCCCAGGTC |
| pcDNAesIFNκ-F | CAGTGTGCTGGAATTCCACCATGAAAACCAAGTCTGATATG |
| pcDNAesIFNκ-R | GATATCTGCAGAATTCTTATTTCCTTCTGAGTAGTTC |
| pcDNAesIFNω-F | CAGTGTGCTGGAATTCCACCATGGCCCTCCTGCTCTC |
| pcDNAesIFNω-R | GATATCTGCAGAATTCTCAAGGTGACCCCAGGTC |
| GAPDH-F | TCGGAGTGAACGGATTTG |
| GAPDH-R | CCTTGAACTTGCCATGAGTAG |
| ISG56-F | CAGGCTAAATCCAGAAGATG |
| ISG56-R | TTCCAGAGCAAATTCAAAAT |
| Mx1-F | TCTACTGCCAAGACCAAGCGT |
| Mx1-R | CGAGGGAGCAAGTCAAAGGA |
| IFIT3-F | AGCAGAGGAGCTTGCAGAAG |
| IFIT3-R | CCGGAAAGCCATAAACAAGA |
| EBLV1-F | GAAAGGKGACAAGATAACACC |
| EBLV1-R | ARAGAAGAAGTCCAACCAGAG |
| EBLV2-F | GGTGTCTGTAAAGCCAGAAG |
| EBLV2-R | TTATAAGCTCTGTTCAAG |
| RABV-F | GATCCTGATGAYGTATGTTCCTA |
| RABV-R | GATTCCGTAGCTRGTCCA |
F, forward primer; R, reverse primer.
In order to obtain the complete gene sequences of esIFN-κ and esIFN-ω, a genome walking method was applied according to the Genome Walker universal kit manual (Clontech, USA). esIFN-κ and esIFN-ω specific genome walking primers were designed based on the gene fragments obtained above (Table 1). For 5′-end walking PCR, gwesIFNκ-R1 and AP1 or gwesIFNω-R1 and AP1 were used in the first-round PCR, and gwesIFNκ-R2 and AP2 or gwesIFNω-R2 and AP2 were used in the second-round PCR (Table 1). For 3′-end walking PCR, gwesIFNκ-F1 and AP1 or gwesIFNω-F1 and AP1 were used in the first-round PCR, and gwesIFNκ-F2 and AP2 or gwesIFNω-F2 and AP2 were used in the second-round PCR (Table 1). For longer promoter sequences, other sets of gene-specific primers, gwesIFNκ-R3/gwesIFNκ-R4 and gwesIFNω-R3/gwesIFNω-R4, were used in an additional round of PCR (Table 1). All of the specific PCR products were cloned and sequenced in both directions.
Sequence and phylogenetic analysis.
The open reading frames (ORFs) of esIFN-κ and esIFN-ω were predicted by the NCBI ORF finder program (http://www.ncbi.nlm.nih.gov/projects/gorf/). The deduced amino acid sequences were BLAST searched against protein databases (http://www.ncbi.nlm.nih.gov/BLAST/) for a homologue search. Signal peptide and protein domains were identified by SMART (http://smart.embl-heidelberg.de/). The transcription start site was predicted with the promoter predictor program (http://www.fruitfly.org/seq_tools/promoter.html). Potential transcription factor binding sites were identified by using the MatInspector program (http://www.genomatix.de). N-glycosylation sites were predicted by PROSITE (http://www.expasy.ch/prosite/). Sequence alignments were performed by the ClustalX program. Phylogenetic trees were constructed by MEGA5.05 using the neighbor-joining method with bootstrap value of n = 1,000. Protein sequences from other species used in the phylogenetic analysis are shown in the legend to Fig. 2.
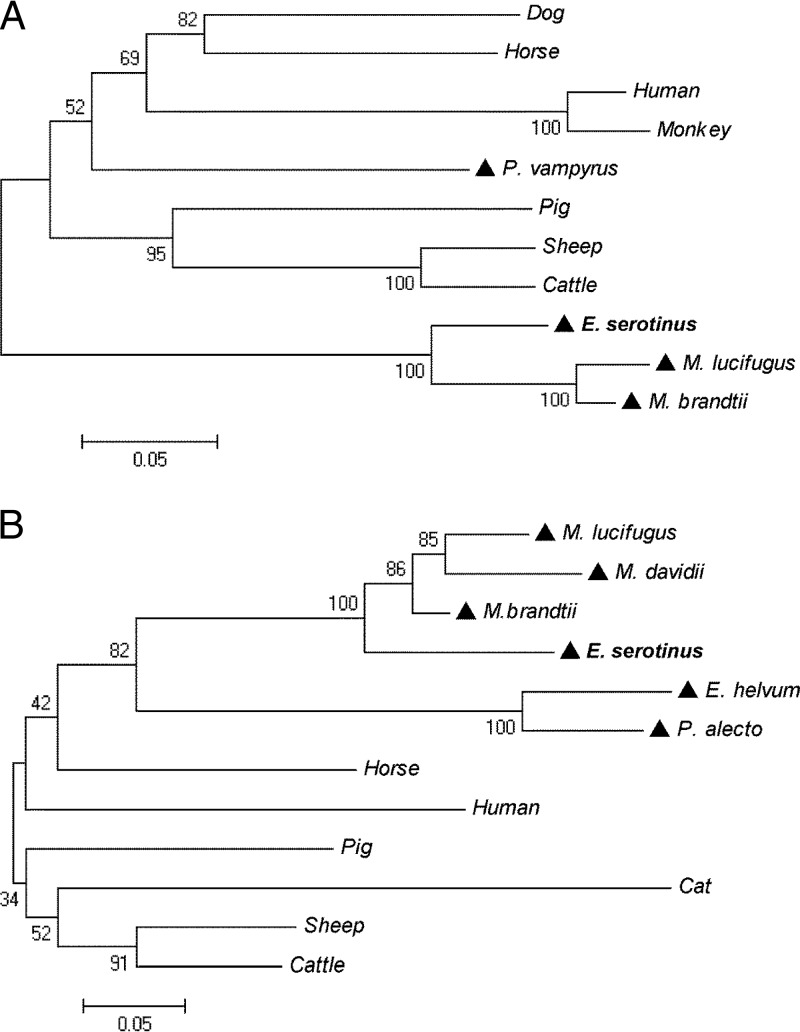
FIG 2.
Phylogenetic analysis of IFN-κ (A) and IFN-ω (B) protein sequences by neighbor-joining method with bootstrap value at n = 1,000. (A) Protein sequences used for IFN-κ: dog, Canis lupus familiaris (XP_003639432); human, Homo sapiens (NP_064509); horse, Equus caballus (XP_001497233); pig, Sus scrofa (NP_001158329); monkey, Nomascus leucogenys (XP_003281871); sheep, Ovis aries (XP_004004472); cattle, Bos Taurus (NP_001193352); and bats, P. vampyrus (ADK60922), M. brandtii (EPQ20610), M. lucifugus (GL430113:491398:492675:1), and E. serotinus (KF758763). (B) Protein sequences used for IFN-ω: human, H. sapiens (EAW58624); horse, E. caballus (XP_003364007); pig, S. scrofa (ACF17563); sheep, O. aries (AAA31507); cattle, B. taurus (XP_876525); cat, Felis catus(NP_001095910); and bats, P. alecto (ELK15819), Eidolon helvum (AFH73816), M. davidii (ELK26495), M. brandtii (EPQ20230), M. lucifugus (GL429988:1422681:1423811:1), and E. serotinus (KF758764). A triangle (▲) indicates a bat species.
Construction of recombinant constructs and expression of recombinant esIFN-κ and esIFN-ω.
An infusion cloning strategy was used to build the recombinant constructs for both prokaryotic and eukaryotic systems. For prokaryotic expression, gene-specific PCR products amplified with the primer pair PmalesIFNκ-F/PmalesIFNκ-R or PmalesIFNω-F/PmalesIFNω-R (Table 1) were inserted into pMAL-c2X plasmids which contain the maltose-binding protein (MBP) tag by using an In-Fusion HD Cloning Plus kit (Clontech). Likewise, for specific eukaryotic constructs, the primer pair pcDNAesIFNκ-F/pcDNAesIFNκ-R or pcDNAesIFNω-F/pcDNAesIFNω-R (Table 1) was used, in which a Kozak sequence was incorporated in the forward primers to favor the initiation of translation (30). PCR products were inserted into pcDNA plasmids by using an In-Fusion HD Cloning Plus kit (Clontech). All of the constructs were confirmed to contain the correct inserts by sequencing. To obtain MBP-tagged recombinant esIFN-κ and esIFN-ω, the pMAL protein fusion and purification system (NEB, United Kingdom) was used. Protein induction and purification analysis were performed as described previously (31). In addition, endotoxins were removed from the purified proteins by using an ToxinEraser endotoxin removal kit (GenScript, USA). The concentration of purified proteins was determined by using NanoDrop.
Cell culture and activity evaluation of esIFN-κ and esIFN-ω.
A naturally immortalized cell line from E. serotinus brain tissue (FLG-R) obtained from the cell culture collection of the Friedrich-Loeffler-Institut (cell bank number FLI-1093) was used to investigate esIFN-κ and esIFN-ω activity. Cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum at 37°C and 5% CO2. The cells were seeded at a density of 4 × 105 cells/well into 12-well tissue culture plates 10 to 12 h before IFN stimulation. After a washing step with 1× phosphate-buffered saline (PBS), the cells were treated with purified MBP-tagged esIFN-κ or esIFN-ω at concentrations of 1 and 10 μg/ml. Cells treated with MBP tag purified from empty pMAL-c2X served as a negative control. Untreated cells were used as blanks. All cells were harvested in RLT buffer (Qiagen) 3 h after stimulation and stored at −80°C for further RNA extraction.
Recombinant IFN from E. coli system without glycosylation may lose its activity; thus, the E. serotinus brain cell line was used for the expression of esIFN-κ and esIFN-ω. Cells were seeded as described above and then transfected with 1.2 μg of pcDNAesIFNκ or pcDNAesIFNω plasmid using 2 μl of Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Transfection with empty plasmid pcDNA3 was used as a negative control, with 10 μg of polyinosinic-polycytidylic acid [poly(I·C); Invivogen]/ml as a positive control and without transfection as a blank control. After transfection and incubation in a humidified atmosphere at 37°C and 5% CO2 for 24 h, the supernatants were harvested and stored at −20°C for an IFN activity test. Cells were collected into RLT buffer and used for RNA extraction. To measure the capability of esIFN-κ and esIFN-ω to activate IFN signaling, the expression level of IFN-stimulated gene 56 (ISG56), myxovirus resistance 1 (Mx1), and IFN-induced protein with tetratricopeptide repeat 3 (IFIT3) as downstream indicators of the pathway were determined by quantitative RT-PCR (qRT-PCR) as described below.
Antiviral assay.
To study the inhibitory effect of IFN-κ and IFN-ω on viral replication, in vitro E. serotinus brain cells were seeded into 24-well plates at a density of 2 × 105 cells/well. At 3 h after seeding, cells were washed with PBS and subsequently stimulated with MBP-tagged esIFN-κ or esIFN-ω at concentrations of 1 and 10 μg/ml for 24 h. Similarly, to test the activity of recombinant esIFN-κ and esIFN-ω overexpressed from the bat cell line, serial dilutions (1:2, 1:8, and 1:32) of the supernatants from transfected cells described above were added instead of MBP-tagged proteins into a final volume of a 500-μl stimulation system. After 24 h, the cells were washed with PBS twice and infected with EBLV-1 (E. serotinus isolate), EBLV-2 (M. daubentoni isolate), or RABV (European fox isolate) at a multiplicity of infection (MOI) of 0.1 for another 24 h. Subsequently, the cells were washed with PBS and collected into RLT, and RNA was prepared by using an RNeasy minikit (Qiagen). The relative viral load was analyzed by qRT-PCR as described below.
qRT-PCR.
The qRT-PCR method was introduced to measure the mRNA expression levels of IFN-κ, IFN-ω, and IFN-induced genes in response to stimulation. The primers used for target genes and internal control GAPDH (glyceraldehyde-3-phosphate dehydrogenase) are listed in Table 1. Likewise, to determine the relative viral loads indicative of virus replication after IFN stimulation in E. serotinus brain cells, qRT-PCR was performed using EBLV-1-, EBLV-2-, and RABV-specific primers to target lyssavirus nucleoprotein genes (Table 1). qRT-PCR was performed using the CFX96 TouchDetection system (Bio-Rad, USA) with a SensiFAST SYBR one-step kit (Bioline, United Kingdom) according to the manufacturer's instructions. To assess the specificity of the PCR amplification, a melting-curve analysis was performed at the end of the reaction. The relative expression levels of targets were calculated by the 2−ΔΔCT method (32).
Statistical analysis.
All data are presented as means ± the standard deviations. Statistically significant differences were analyzed by one-way analysis of variance using the SPSS software package.
Nucleotide sequence accession numbers.
Complete ORFs and partial promoter regions were submitted to GenBank under accession numbers KF758763 (esIFN-κ) and KF758764 (esIFN-ω).
RESULTS
Sequence characteristics of esIFN-κ and esIFN-ω.
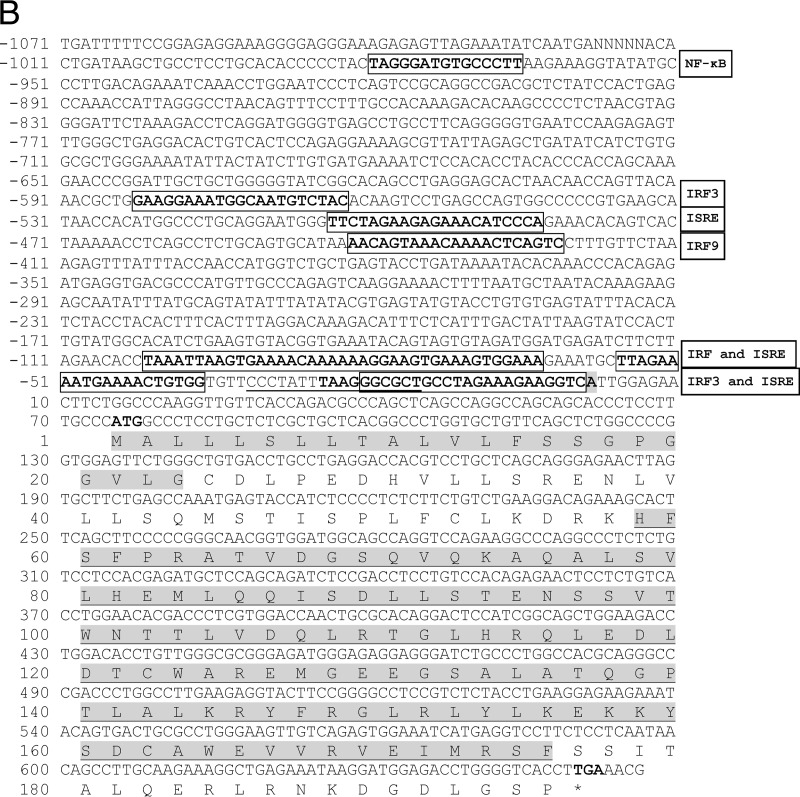
IFN-κ (scaffold no. GL430113; bp 491398 to 492675) and IFN-ω (scaffold no. GL429988; bp 1422681 to 1423811) genes of M. lucifugus in Ensembl were used as reference sequences to design primers to amplify 376 bp of esIFN-κ and 404 bp of esIFN-ω specific fragments from E. serotinus. Complete ORFs and partial promoter regions were further obtained by Genome walking and submitted to GenBank. ORFs of esIFN-κ and esIFN-ω consisted of 627 bp (encoding 208 amino acids) and 588 bp (encoding 195 amino acids), respectively (Fig. 1). Both IFNs contain a signal peptide and interferon alpha, beta, and delta domains (IFabd) (Fig. 1). Sequence analysis revealed that both esIFN-κ and esIFN-ω genes are intronless. The potential transcription start sites of both genes were indicated by a promoter predictor revealing 2,038 bp for esIFN-κ and 1,071 bp for esIFN-ω promoter sequences (Fig. 1). Transcription factor binding sites such as IRFs, ISREs, and NF-κB were found in both promoter regions by MatInspector (Fig. 1). N-glycosylation sites were identified in esIFN-κ (50-NMSK-53) and esIFN-ω (95-NSSV-98) by the PROSITE tool (Fig. 1).
FIG 1.
The genomic sequence and deduced amino acid sequences of esIFN-κ (A) and esIFN-ω (B) genes. The start and stop codons are shown in bold. The signal peptide is in gray. The IFabd domain is shaded gray and underlined. The predicted transcription start site is shaded gray and bold. The TATA box element is underlined. and core sequence is in boldface. Some of the predicted transcription factor binding sites are in bold and boxed.
Phylogenetic analysis.
Phylogenetic trees of IFN-κ and IFN-ω were constructed based on alignments of protein sequences from E. serotinus with sequences from 12 mammal species, including one human and five Chiroptera, by the neighbor-joining method. The results showed that the IFNs of bats are distantly related to those of other mammals and humans, with Megachiroptera and Microchirotpera obviously forming two separate genetic groups (Fig. 2). By sharing 87 to 95% protein sequence similarities, IFN-κs from E. serotinus, M. lucifugus, and M. brandtii formed a sister branch. In contrast, IFN-κ from the megabat P. vampyrus clustered first into a nonbat mammalian group. The selected IFN-ω protein sequences of different bat species share 59 to 90% sequence similarities and were grouped together.
Recombinant esIFN-κ and esIFN-ω initiate IFN signaling in the E. serotinus brain cell line.
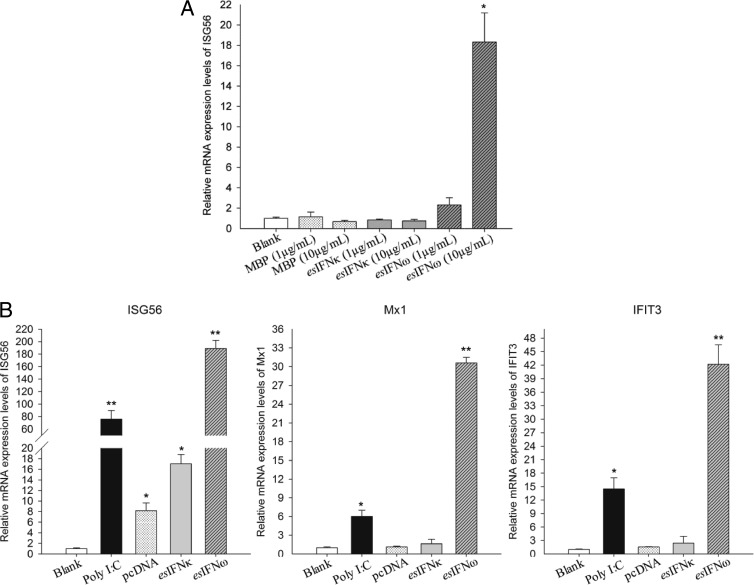
To evaluate the biological activities of both IFNs, the expression levels of ISG56 mRNA were assessed by qRT-PCR after activation by E. coli-derived recombinant esIFN-κ and esIFN-ω. In esIFN-ω-treated cells, ISG56 expression appeared to be dose dependent, with increases of 2-fold in the 1-μg/ml group and >17-fold in the 10-μg/ml group (P < 0.05) (Fig. 3A). In contrast to esIFN-ω stimulation, ISG56 expression was not induced by esIFN-κ (Fig. 3A).
FIG 3.
esIFN-κ and esIFN-ω initiate IFN signaling in an E. serotinus brain cell line. (A) Cells were treated with either recombinant esIFN-κ and esIFN-ω MBP-tagged proteins (1 and 10 μg/ml) or MBP alone (1 and 10 μg/ml) for 3 h as described in the text. Only the higher concentration (10 μg/ml) of recombinant esIFN-ω induced a significant increase in ISG56 mRNA (*, P < 0.05), as measured by qRT-PCR. (B) Cells were transfected with esIFN-κ and esIFN-ω expression plasmids (1.2 μg/well in a 12-well plate) or poly(I·C) at 10 μg/ml as a positive control. Note that eukaryotic expressed esIFN-κ and esIFN-ω induced a much stronger ISG56 mRNA level, as measured by qRT-PCR. The expressions of Mx1 and IFIT3 were also upregulated by esIFN-ω.
To ensure that the expression in E. coli does not influence biological activity, esIFN-κ and esIFN-ω were also expressed in the E. serotinus cell line. The qRT-PCR results showed that both recombinant esIFN-κ and recombinant esIFN-ω can induce the expression of ISG56 (Fig. 3B). In particular, in pcDNAesIFNω-transfected cells ISG56 expression was 100-fold higher than in the positive control transfected with poly(I·C) (P < 0.01) (Fig. 3B). The upregulation of ISG56 in pcDNAesIFNκ-transfected cells induced a 9-fold enhancement compared to empty plasmid (P < 0.05) (Fig. 3B). In addition to ISG56, Mx1 and IFIT3 were also elevated 5- and 14-fold, respectively, in response to poly(I·C) stimulation (P < 0.05) and 30- and 41-fold, respectively, in response to pcDNAesIFNω transfection (P < 0.01) (Fig. 3B).
esIFN-κ and esIFN-ω inhibit the replication of lyssaviruses in a bat cell line.
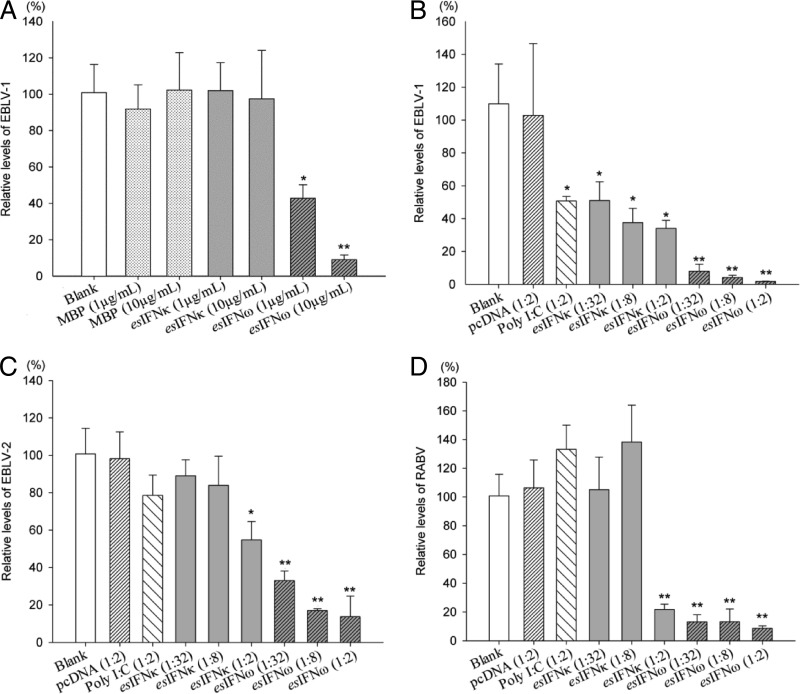
The inhibition of lyssavirus replication was determined by qRT-PCR to measure the relative virus RNA levels after infection in esIFN-κ- and esIFN-ω-pretreated cell lines. Generally, both esIFN-κ and esIFN-ω can inhibit lyssavirus EBLV-1, EBLV-2, and RABV replication in the E. serotinus brain cell line to different extents. Although esIFN-κ did not influence the viral replication of EBLV-1, esIFN-ω showed a dose-dependent suppression (58 to 91% reduction) (P < 0.05) (Fig. 4A). The control tag MBP did not inhibit EBLV-1 replication (Fig. 4A).
FIG 4.
esIFN-κ and esIFN-ω inhibit lyssavirus replication in an E. serotinus brain cell line. (A) Cells pretreated for 24 h with recombinant E. coli-derived esIFN-κ and esIFN-ω MBP-tagged proteins (1 and 10 μg/ml) or MBP alone (1 and 10 μg/ml) and then infected with EBLV-1 (MOI of 0.1) for 24 h. (B to D) Cells pretreated 24 h with recombinant eukaryotic expressed IFN in supernatants from an esIFN-κ- and esIFN-ω-transfected E. serotinus brain cell line and infected with EBLV-1 (B), EBLV-2 (C), or RABV (D) (MOI of 0.1) for 24 h. The viral replication was determined by qRT-PCR for lyssavirus nucleoprotein gene. Only the E. coli-derived esIFN-ω MBP-tagged proteins inhibited the EBLV-1 replication in a dose-dependent manner. In contrast, both recombinant eukaryotic derived esIFN-κ and esIFN-ω inhibited the replication of EBLV-1 (B), EBLV-2 (C), or RABV (D) in a dose-dependent manner. However, esIFN-κ showed a much weaker activity, especially against EBLV-2 and RABV.
In further step, both esIFN-κ- and esIFN-ω-containing supernatants were used for the antiviral assays against EBLV-2 and RABV, in addition to EBLV-1. In the EBLV-1-infected group, supernatants from poly(I·C)- and pcDNAesIFNκ-transfected cells showed moderately inhibitory effects (54 to 69% reduction) on EBLV-1 replication (P < 0.05), whereas the empty plasmid pcDNA3 supernatant showed no effect (Fig. 4B). The esIFN-ω supernatant led to a dose-dependent reduction of virus RNA from 92 to 98% (P < 0.01) (Fig. 4B).
In the EBLV-2-infected group, poly(I·C)-transfected supernatants showed a slight inhibitory potency to EBLV-2 infection (Fig. 4C). Although esIFN-κ reduced 11 to 46% of the virus RNA at different concentrations, esIFN-ω decreased 67 to 86% (P < 0.01) (Fig. 4C). In the RABV-infected group, the negative control and poly(I·C)-transfected supernatants had no effect on RABV infection (Fig. 4D). Suppression effects were observed in the esIFN-κ-treated groups at high concentrations (78% reduction) and in the esIFN-ω-treated groups at all concentrations used (87 to 91%) (P < 0.01; Fig. 4D). Overall, both recombinant esIFN-κ and esIFN-ω showed antiviral properties in the E. serotinus brain cell line, but esIFN-ω had a stronger effect than esIFN-κ. The inhibitory effect of esIFN-ω against different lyssaviruses varies, with the order EBLV-1 > RABV > EBLV-2.
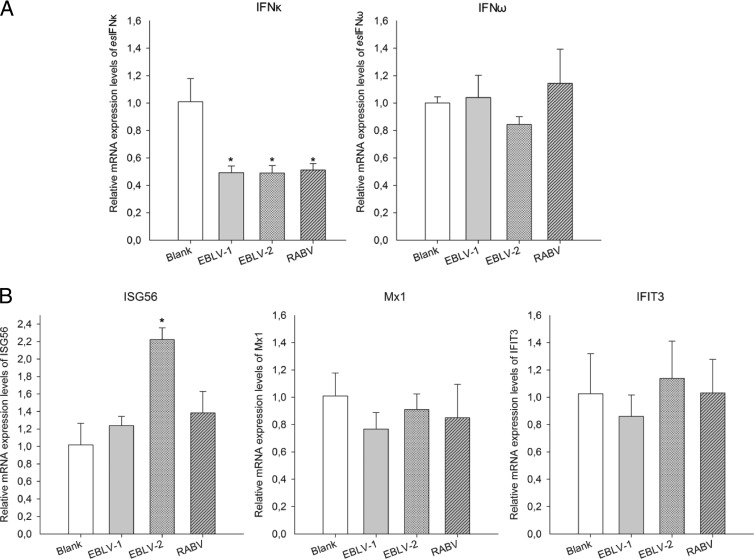
Expression patterns of IFNs and IFN-induced genes during lyssavirus infection.
To investigate the interaction between host IFN signaling and lyssavirus, the mRNA expression of esIFN-κ, esIFN-ω, and the IFN-induced genes ISG56, Mx1, and IFIT3 was measured by qRT-PCR after lyssavirus infection. Overall, a silent expression pattern was observed (Fig. 4A and B). IFN-ω expression did not change during infection, whereas IFN-κ was actually downregulated 50% by all three viruses (P < 0.01) (Fig. 5A). Similarly, the three tested IFN-induced genes remained silent or at a nearly silent level with the exception of ISG56, which slightly increased in response to EBLV-2 infection (P < 0.05) (Fig. 5B).
FIG 5.
Expression patterns of esIFN-κ, esIFN-ω, and IFN-induced genes after lyssavirus infection. An E. serotinus brain cell line was infected with EBLV-1, EBLV-2, or RABV (MOI of 0.1) for 24 h, and the expression of esIFN-κ and esIFN-ω (A) and IFN-induced genes ISG56, Mx1, and IFIT3 (B) was measured by qRT-PCR. Note the downregulation of esIFN-κ by 50% in the virus-infected cells (A) and the weakly increased expression of ISG56 after EBLV-2 infection, whereas the expression level of all of the other IFN-induced genes was not changed (B).
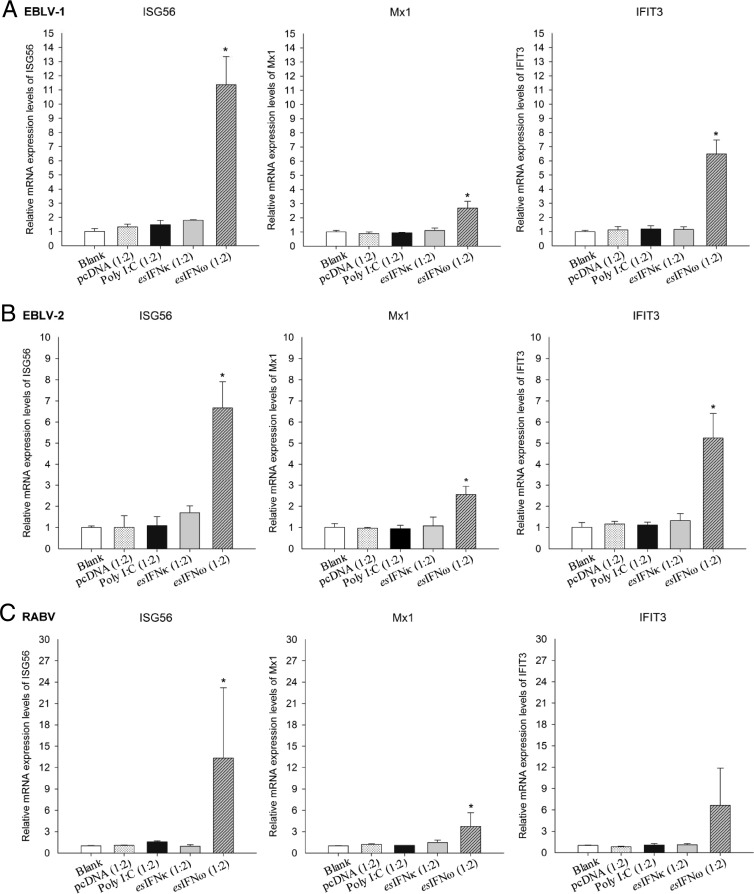
To further evaluate whether the esIFN-κ- and esIFN-ω-mediated antiviral responses were still induced efficiently after viral infection, the expression levels of IFN-induced genes were quantified. ISG56, Mx1, and IFIT3 expression displayed a variously upregulated level in IFN-ω-pretreated, subsequently infected cells (P < 0.05) (Fig. 6). In contrast, no induced expression was seen in the IFN-κ-pretreated, subsequently infected cells (Fig. 6).
FIG 6.
Expression patterns of IFN induced genes in esIFN-κ- and esIFN-ω-pretreated cells after lyssavirus infection. The cells were pretreated with recombinant eukaryotic expressed IFN in supernatants from esIFN-κ- and esIFN-ω-transfected E. serotinus brain cell line and then infected with EBLV-1 (A), EBLV-2 (B), and RABV (C) (MOI of 0.1) for 24 h. The expression of IFN-induced genes ISG56, Mx1, and IFIT3 was determined by qRT-PCR. Only in IFN-ω-pretreated cells did ISG56, Mx1, and IFIT3 expression displayed a low upregulation, whereas in IFN-κ-pretreated cells, no induced induction of these genes was seen.
DISCUSSION
The identification of bats as a potential reservoir for zoonotic diseases, such as those caused by RABV, SARS-CoV, or Hendra virus, has drawn attention to the bat immune system (6). In particular, the ability of bats to host these infections without obvious clinical signs is still enigmatic to the scientific community (33, 34). Presumably, the long coevolution contributed to the development of protective mechanisms, allowing a long coexistence of bats together with their viruses. As in other species, IFNs play a pivotal role in the antiviral protection of the host. To address this issue for European bats in the context of lyssavirus infections, we studied the activities of IFNs in vitro using established an bat cell line.
Thus, we focused here on cloning and characterizing type I IFNs from E. serotinus. First, IFN-κ and -ω were cloned completely, whereas IFN-α and -β sequences could not be found. Therefore, we concentrated on molecular and functional characterization of IFN-κ and -ω from the serotine bat. The sequence characteristics of intronless genomic organization, conserved N-glycosylation sites of esIFN-κ and esIFN-ω demonstrated the structural similarities of these two type I IFNs from serotine bat to those of other mammals (35). Furthermore, these findings indicate that bat IFN-κ and IFN-ω probably share similar functions with the IFNs of other mammals; this was also confirmed by structural conservation of the esIFN-κ and esIFN-ω promoter region. Transcription factors such as IRF, ISRE, ATF2/c-Jun, and NF-κB binding sites could be identified in the proximal promoter regions of esIFN-κ and esIFN-ω, suggesting that the transcriptional regulation of bat type I IFNs is fundamentally similar to that of other mammals. However, the different IRF members and various positions of IRF and NF-κB binding elements suggested that esIFN-κ and esIFN-ω are likely to be regulated in a different manner. The identification of transcription factor binding sites is just the first step in investigating the intricate regulatory network of the less-studied type I IFN members κ and ω. To address the interactions between these regulatory elements and transcription factors and to understand how the transcription factors cooperate to regulate the expression of IFN-κ and IFN-ω, functional studies are required.
All type I IFN genes have been identified in mammals in one single chromosomal region and expanded by duplication during evolution (17, 36). However, the IFN-κ gene is found outside the type I IFN locus, suggesting an independent development in different mammalian families (36). Given the evolutionary relatedness of bats (37, 38), IFN-κ gene sequences in bat species should be more diversified. In fact, the results of our phylogenetic analysis, in which IFN-κ sequences from microchiroptera and megachiroptera grouped separately from other mammals (including humans), support this hypothesis. Interestingly, both IFNs of E. serotinus grouped with other microbat-associated sequences, albeit a clear separation from Myotis IFNs—i.e., M. lucifugus, M. brandtii, and M. davidii—is visible. A similar genetic separation between these two bat families was also observed following comparison of the cytochrome b sequences used for species identification (39). The sequence conservation, as well as the diversification of IFN-κ and IFN-ω in different bat families, implies the presence of evolving and adaptable IFNs in bat species.
To determine the functional characteristics of IFN-κ and IFN-ω in bats against virus infection, recombinant IFN-κ and IFN-ω from E. serotinus were expressed in both prokaryotic and eukaryotic systems and used for functional studies. The upregulation of IFN-induced genes by esIFN-κ and esIFN-ω demonstrated that both IFNs work as functional molecules, inducing IFN signal pathways in bat cell lines. Particularly, different properties of esIFN-κ and esIFN-ω in the activation of IFN-induced genes implied their functional divergence in downstream signaling or effecting cell types.
To further study their antiviral functions, the activities of esIFN-κ and esIFN-ω were investigated against different lyssaviruses (EBLV-1, EBLV-2, and RABV) in an E. serotinus brain cell line. Both esIFN-κ and esIFN-ω proteins demonstrated antiviral activity. This is consistent with type I IFNs limiting RABV infection in mice (12). Previous studies reported that recombinant type III IFN and type II IFN from P. alecto can inhibit Pulau virus replication and Semliki Forest virus and Hendra virus replication, respectively (22, 23). However, the antiviral activity of bat type I IFNs is less studied. In addition, no study has yet shown whether bat IFNs are pathogen associated. In the present study, the different inhibitory extents of esIFN-κ and esIFN-ω to EBLV-1, EBLV-2, and RABV provided evidence for such an association between IFNs and different viruses. The strong inhibition of EBLV-1 replication in the E. serotinus cell line by these two type I IFNs suggests that esIFN-κ and esIFN-ω are involved in an anti-lyssavirus innate immune defense mechanisms in natural hosts.
However, as shown by the experiments performed here, the interaction between the virus and the bat IFN response seems to be more complex. Although the restriction of viral replication in the IFN pretreated cells suggested a robust IFN-mediated innate immune reaction limiting virus spread at an early infection stage, the general silencing of esIFN-κ, esIFN-ω, and their induced genes during infection indicates that bat-associated lyssaviruses can interfere with the signaling cascades, leading to transcriptional activation of the IFN system. This further highlights the importance of lyssaviral countermeasures against the host type I IFN system for the establishment of an infection (14, 40, 41). Taken together, these results indicate a balance of persistent infection and limited viral growth, elucidating our understanding of the coexistence of lyssaviruses in bats under natural conditions.
To conclude, type I IFN-κ and IFN-ω were cloned and functionally characterized from E. serotinus. Sequence and functional analysis revealed that whereas type I IFNs are generally conserved among mammals, these IFNs are evolutionally divergent and adapted in bats. Analysis of the promoter regions of esIFN-κ and esIFN-ω provided evidence for the control architecture of IFN-κ and IFN-ω transcription, and functional studies indicated their different capacities in antiviral activity. Overall, the present study of esIFN-κ and esIFN-ω provides functional data on the interactions of the IFN system and lyssaviruses in bats, add to our understanding of the long-term coexistence of bats with rabies, and expands our general knowledge of type I IFNs in mammals. In an ongoing functional study, we are investigating type I IFNs, including IFN-α, -β, -κ, and -ω, in order to understand the complex interactions between lyssaviruses and their natural host in vitro in a more comprehensive way.
ACKNOWLEDGMENTS
This study was financially supported by the Bundesministerium für Bildung und Forschung (grant 01KI1016A).
We thank Stefan Finke for the RABV strain, Günther Keil for the pcDNA3 plasmid, and Yang Zhang for the pMAL-c2X plasmid.
Footnotes
Published ahead of print 26 February 2014
REFERENCES
- 1.Altringham JD. 2011. Bats: from evolution to conservation, 2nd ed, p 319 Oxford University Press, Inc, New York, NY [Google Scholar]
- 2.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. 2006. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 19:531–545. 10.1128/CMR.00017-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang LF. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679. 10.1126/science.1118391 [DOI] [PubMed] [Google Scholar]
- 4.Halpin K, Young PL, Field HE, Mackenzie JS. 2000. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J. Gen. Virol. 81:1927–1932 [DOI] [PubMed] [Google Scholar]
- 5.Muller T, Cox J, Peter W, Schafer R, Johnson N, McElhinney LM, Geue JL, Tjornehoj K, Fooks AR. 2004. Spill-over of European bat lyssavirus type 1 into a stone marten (Martes foina) in Germany. J. Vet. Med. B Infect. Dis. Vet. Public Health 51:49–54. 10.1111/j.1439-0450.2003.00725.x [DOI] [PubMed] [Google Scholar]
- 6.Wibbelt G, Moore MS, Schountz T, Voigt CC. 2010. Emerging diseases in Chiroptera: why bats? Biol. Lett. 6:438–440. 10.1098/rsbl.2010.0267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris SL, Brookes SM, Jones G, Hutson AM, Fooks AR. 2006. Passive surveillance (1987 to 2004) of United Kingdom bats for European bat lyssaviruses. Vet. Rec. 159:439–446. 10.1136/vr.159.14.439 [DOI] [PubMed] [Google Scholar]
- 8.Baker ML, Schountz T, Wang LF. 2013. Antiviral immune responses of bats: a review. Zoonoses Public Health 60:104–116. 10.1111/j.1863-2378.2012.01528.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobson AP. 2005. Virology: what links bats to emerging infectious diseases? Science 310:628–629. 10.1126/science.1120872 [DOI] [PubMed] [Google Scholar]
- 10.Paul BN, Chakravarty AK. 1986. In vitro analysis of delayed immune response in a bat, Pteropus giganteus: process of con-A mediated activation. Dev. Comp. Immunol. 10:55–67. 10.1016/0145-305X(86)90044-3 [DOI] [PubMed] [Google Scholar]
- 11.Chakravarty AK, Sarkar SK. 1994. Immunofluorescence analysis of immunoglobulin bearing lymphocytes in the Indian fruit bat: Pteropus giganteus. Lymphology 27:97–104 [PubMed] [Google Scholar]
- 12.Chopy D, Detje CN, Lafage M, Kalinke U, Lafon M. 2011. The type I interferon response bridles rabies virus infection and reduces pathogenicity. J. Neurovirol. 17:353–367. 10.1007/s13365-011-0041-6 [DOI] [PubMed] [Google Scholar]
- 13.Chopy D, Pothlichet J, Lafage M, Megret F, Fiette L, Si-Tahar Lafon MM. 2011. Ambivalent role of the innate immune response in rabies virus pathogenesis. J. Virol. 85:6657–6668. 10.1128/JVI.00302-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rieder M, Conzelmann KK. 2011. Interferon in rabies virus infection. Adv. Virus Res. 79:91–114. 10.1016/B978-0-12-387040-7.00006-8 [DOI] [PubMed] [Google Scholar]
- 15.Rieder M, Finke S, Conzelmann KK. 2012. Interferon in lyssavirus infection. Berl. Munch. Tierarztl. Wochenschr. 125:209–218 [PubMed] [Google Scholar]
- 16.Walker AM, Roberts RM. 2009. Characterization of the bovine type I IFN locus: rearrangements, expansions, and novel subfamilies. BMC Genomics 10:187. 10.1186/1471-2164-10-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz MO, Pomykala HM, Bohlander SK, Maltepe E, Malik K, Brownstein B, Olopade OI. 1994. Structure of the human type-I interferon gene cluster determined from a YAC clone contig. Genomics 22:540–552. 10.1006/geno.1994.1427 [DOI] [PubMed] [Google Scholar]
- 18.LaFleur DW, Nardelli B, Tsareva T, Mather D, Feng P, Semenuk M, Taylor K, Buergin M, Chinchilla D, Roshke V, Chen G, Ruben SM, Pitha PM, Coleman TA, Moore PA. 2001. Interferon-kappa, a novel type I interferon expressed in human keratinocytes. J. Biol. Chem. 276:39765–39771. 10.1074/jbc.M102502200 [DOI] [PubMed] [Google Scholar]
- 19.Zhou P, Cowled C, Wang LF, Baker ML. 2013. Bat Mx1 and Oas1, but not Pkr, are highly induced by bat interferon and viral infection. Dev. Comp. Immunol. 40:240–247. 10.1016/j.dci.2013.03.006 [DOI] [PubMed] [Google Scholar]
- 20.Biesold SE, Ritz D, Gloza-Rausch F, Wollny R, Drexler JF, Corman VM, Kalko EK, Oppong S, Drosten C, Muller MA. 2011. Type I interferon reaction to viral infection in interferon-competent, immortalized cell lines from the African fruit bat Eidolon helvum. PLoS One 6:e28131. 10.1371/journal.pone.0028131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Virtue ER, Marsh GA, Baker ML, Wang LF. 2011. Interferon production and signaling pathways are antagonized during henipavirus infection of fruit bat cell lines. PLoS One 6:e22488. 10.1371/journal.pone.0022488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janardhana V, Tachedjian M, Crameri G, Cowled C, Wang LF, Baker ML. 2012. Cloning, expression, and antiviral activity of IFNγ from the Australian fruit bat, Pteropus alecto. Dev. Comp. Immunol. 36:610–618. 10.1016/j.dci.2011.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou P, Cowled C, Todd S, Crameri G, Virtue ER, Marsh GA, Klein R, Shi Z, Wang LF, Baker ML. 2011. Type III IFNs in pteropid bats: differential expression patterns provide evidence for distinct roles in antiviral immunity. J. Immunol. 186:3138–3147. 10.4049/jimmunol.1003115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Omatsu T, Bak EJ, Ishii Y, Kyuwa S, Tohya Y, Akashi H, Yoshikawa Y. 2008. Induction and sequencing of Rousette bat interferon alpha and beta genes. Vet. Immunol. Immunopathol. 124:169–176. 10.1016/j.vetimm.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amengual B, Whitby JE, King A, Cobo JS, Bourhy H. 1997. Evolution of European bat lyssaviruses. J. Gen. Virol. 78(Pt 9):2319–2328 [DOI] [PubMed] [Google Scholar]
- 26.Kuzmin IV, Hughes GJ, Botvinkin AD, Orciari LA, Rupprecht CE. 2005. Phylogenetic relationships of Irkut and West Caucasian bat viruses within the Lyssavirus genus and suggested quantitative criteria based on the N gene sequence for lyssavirus genotype definition. Virus Res. 111:28–43. 10.1016/j.virusres.2005.03.008 [DOI] [PubMed] [Google Scholar]
- 27.Freuling CM, Beer M, Conraths FJ, Finke S, Hoffmann B, Keller B, Kliemt J, Mettenleiter TC, Muhlbach E, Teifke JP, Wohlsein P, Muller T. 2011. Novel lyssavirus in Natterer's bat, Germany. Emerg. Infect. Dis. 17:1519–1522. 10.3201/eid1708.110201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arechiga Ceballos N, Vazquez Moron S, Berciano JM, Nicolas O, Aznar Lopez C, Juste J, Rodriguez Nevado C, Aguilar Setien A, Echevarria JE. 2013. Novel lyssavirus in bat, Spain. Emerg. Infect. Dis. 19:793–795. 10.3201/eid1905.121071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schatz J, Fooks AR, McElhinney L, Horton D, Echevarria J, Vazquez-Moron S, Kooi EA, Rasmussen TB, Müller T, Freuling CM. 2013. Bat rabies surveillance in Europe. Zoonoses Public Health 60:22–34. 10.1111/zph.12002 [DOI] [PubMed] [Google Scholar]
- 30.De Angioletti M, Lacerra G, Sabato V, Carestia C. 2004. Beta+45 G→C: a novel silent beta-thalassaemia mutation, the first in the Kozak sequence. Br. J. Haematol. 124:224–231. 10.1046/j.1365-2141.2003.04754.x [DOI] [PubMed] [Google Scholar]
- 31.He X, Zhang Y, Yu Z. 2011. An Mpeg (macrophage expressed gene) from the Pacific oyster Crassostrea gigas: molecular characterization and gene expression. Fish Shellfish Immunol. 30:870–876. 10.1016/j.fsi.2011.01.009 [DOI] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 33.Freuling C, Vos A, Johnson N, Fooks AR, Muller T. 2009. Bat rabies: a Gordian knot? Berl. Munch. Tierarztl. Wochenschr. 122:425–433 [PubMed] [Google Scholar]
- 34.Wynne JW, Wang LF. 2013. Bats and viruses: friend or foe? PLoS Pathog. 9:e1003651. 10.1371/journal.ppat.1003651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hughes AL. 1995. The evolution of the type I interferon gene family in mammals. J. Mol. Evol. 41:539–548 [DOI] [PubMed] [Google Scholar]
- 36.Hardy MP, Owczarek CM, Jermiin LS, Ejdeback M, Hertzog PJ. 2004. Characterization of the type I interferon locus and identification of novel genes. Genomics 84:331–345. 10.1016/j.ygeno.2004.03.003 [DOI] [PubMed] [Google Scholar]
- 37.Pettigrew JD, Jamieson BG, Robson SK, Hall LS, McAnally KI, Cooper HM. 1989. Phylogenetic relations between microbats, megabats, and primates (Mammalia: Chiroptera and Primates). Philos. Trans. R. Soc. Lond. B Biol. Sci. 325:489–559. 10.1098/rstb.1989.0102 [DOI] [PubMed] [Google Scholar]
- 38.Mindell DP, Dick CW, Baker RJ. 1991. Phylogenetic relationships among megabats, microbats, and primates. Proc. Natl. Acad. Sci. U. S. A. 88:10322–10326. 10.1073/pnas.88.22.10322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruedi M, Stadelmann B, Gager Y, Douzery EJ, Francis CM, Lin LK, Guillen-Servent A, Cibois A. 2013. Molecular phylogenetic reconstructions identify East Asia as the cradle for the evolution of the cosmopolitan genus Myotis (Mammalia, Chiroptera). Mol. Phylogenet. Evol. 69:437–449. 10.1016/j.ympev.2013.08.011 [DOI] [PubMed] [Google Scholar]
- 40.Wiltzer L, Larrous F, Oksayan S, Ito N, Marsh GA, Wang L, Blondel D, Bourhy H, Jans DA, Moseley GW. 2012. Conservation of a unique mechanism of immune evasion across the Lyssavirus genus. J. Virol. 86:10194–10199. 10.1128/JVI.01249-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lieu KG, Brice A, Wiltzer L, Hirst B, Jans DA, Blondel D, Moseley GW. 2013. The rabies virus interferon antagonist P protein interacts with activated STAT3 and inhibits Gp130 receptor signaling. J. Virol. 87:8261–8265. 10.1128/JVI.00989-13 [DOI] [PMC free article] [PubMed] [Google Scholar]