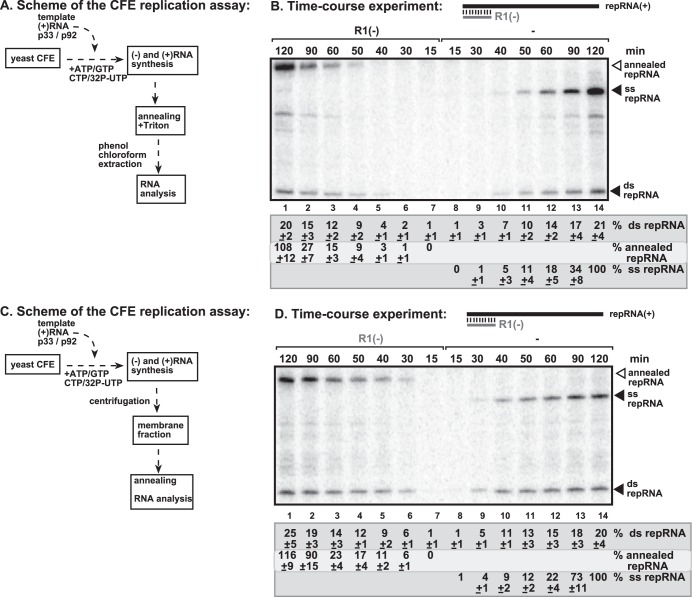
FIG 4.
Time course to detect the appearance of TBSV repRNA products generated in the CFE assay. (A) Scheme of the CFE-based TBSV replication assay. At the end of the replication assay, we added unlabeled R1(−) RNA to the CFE assay mixture to test the polarity of the ssRNA products. (B) Representative nondenaturing PAGE analysis of 32P-labeled repRNA products synthesized by the tombusvirus replicase in the CFE assay. The positions of shifted repRNAs [due to the annealing to R1(−) RNA], single-stranded repRNAs, and double-stranded repRNAs are shown. Note that the production of dsRNA and ssRNA products requires a longer time in this assay than in the assay depicted in Fig. 1 because here we did not preassemble the replicase complex prior to the addition of ribonucleotides. Each experiment was repeated three times. (C) Scheme of the CFE-based TBSV replication assay. At the end of the time course of the replication assay, we removed the supernatant by centrifugation and added unlabeled R1(−) RNA (in the presence of 0.1% Triton X-100) to the membrane fraction of the CFE to test the polarity of the 32P-labeled ssRNA products. Note that only the membrane-bound 32P-labeled repRNA products were analyzed in order to get rid of the large amount of original (+)repRNA transcripts added at the start of the CFE assay. (D) Representative PAGE analysis of 32P-labeled repRNA products synthesized by the tombusvirus replicase in the CFE assay. The positions of shifted repRNAs [due to the annealing to R1(−) RNA], single-stranded repRNAs, and double-stranded repRNAs are shown. Note that the bulk amount of dsRNA appeared earlier (between 30 and 50 min) than that of the (+)ssRNA product (between 60 and 120 min), suggesting that the dsRNA might serve as a template for new (+)RNA synthesis and less likely that the dsRNA is a dead-end product of replication. Each experiment was repeated three times.