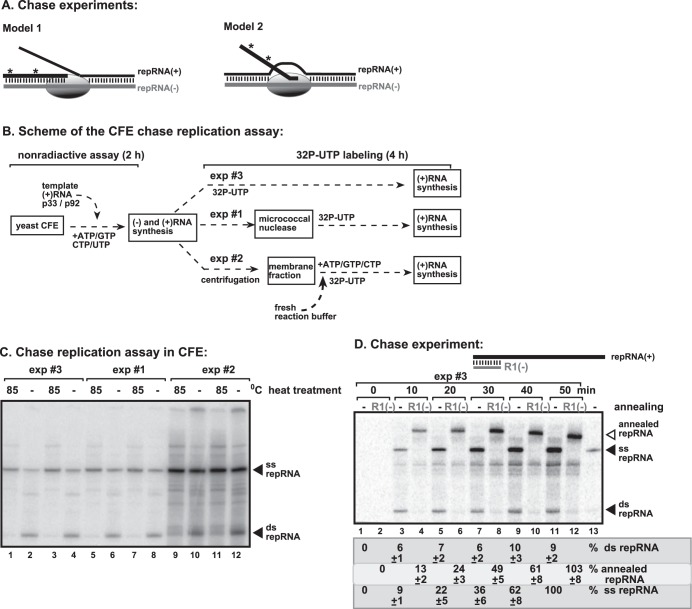
FIG 5.
TBSV replicates via a strand displacement mechanism during plus-strand synthesis. (A) Semiconservative (strand displacement) (model 1) and conservative (model 2) models of (+)RNA synthesis based on dsRNA templates. Note the different fate of the newly synthesized 32P-labeled (+)RNA depending on the mechanism of replication. (B) Schemes of the 32P-labeled UTP chase experiments. In experiment 1, micrococcal nuclease treatment (lasting for 15 min followed by inactivation) was applied prior to the addition of 32P-labeled UTP. In experiment 2, the membrane fraction from the prior CFE assay was collected to remove unincorporated nonlabeled ribonucleotides, followed by the addition of new buffer and a new batch of ribonucleotides, including 32P-labeled UTP. The reaction assay was then continued for another 4 h. In experiment 3, 32P-labeled UTP was added to the CFE-based replication assay mixture at the 2-h time point, followed by an additional 4-h reaction. Note that nuclease treatment or centrifugation in experiments 1 and 2 was done to prevent the synthesis of new minus-stranded RNAs in the CFE after the addition of 32P-labeled UTP. (C) PAGE analysis of the 32P-labeled TBSV repRNA products obtained in the CFE assays described above for panel B. The even-numbered lanes represent replicase products without heat treatment (thus, both ssRNA and dsRNA products are visible), while the odd-numbered lanes show the heat-treated replicase products (only ssRNA is present) (for further details, see Fig. 1). (D) RNA shift assay based on annealing of short unlabeled cRNA in chase time course studies. The assay was performed as described above for the experiment 3 scheme in panel B, except that the reaction was stopped after 10 to 50 min, as shown, and after heat denaturation, short R1(−) RNA complementary to the 5′ end of (+)repRNA was added to every second sample (even-numbered rows) to form a partial duplex. The PAGE gel shows the complete shift of the newly synthesized repRNAs (derived from both ssRNA and dsRNA), demonstrating that the new 32P-labeled RNAs represented (+)repRNAs, while the level of 32P-labeled (−)RNA was below the detection limit.