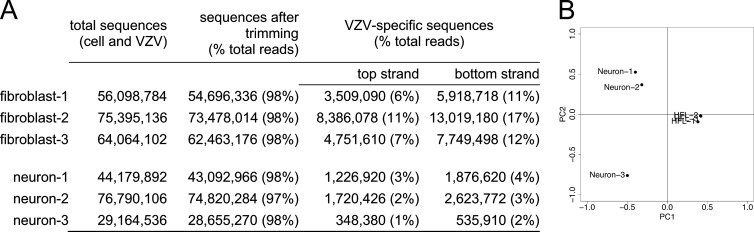
FIG 1.
Analysis of NextGen RNA-seq data quality. (A) Quantification of viral transcripts identified by RNA-seq from VZV-infected fibroblasts and human neurons. (B) Variance among all six samples was analyzed. FPKMs from all libraries were separated using principal-component analysis (PCA). Principal component 1 (PC1) separated samples by their largest variance and resulted in separation between cell types. PC2 separated samples by the next-largest variance, independent of the first, and resulted in separation of samples within cell types. PCA analysis indicated a lack of variation between all three fibroblast samples, whereas neuron 3 was different from neurons 1 and 2. PCA analysis showed that neuron 3 was an outlier.